Figure 4: Defective single-molecule FRET signature of G156E-FUS is ineffectively recovered by WT FUS.
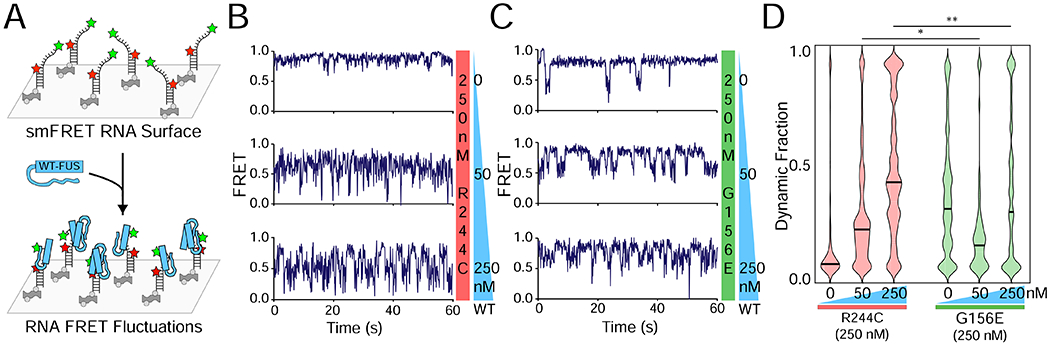
(A) Schematic of FUS binding to single RNAs in smFRET measurements. (B) Sample FRET efficiency traces for FRET-labeled pdU50 RNA with 250 nM R244C and increasing WT concentrations (0 nM, 50 nM, and 250 nM). (C) Same as (B) but with G156E. (D) Violin plot of the dynamic fraction of >100 traces for each condition in (B) and (C). The dynamic fraction represents the proportion of the trace with rapid FRET fluctuations (see Methods). The black bar indicates the median dynamic fraction for each condition. Significance was calculated using a two-tailed two-sample Student’s t-test with ns = not significant, * = p < 0.05, and ** = p < 0.01. Degrees of freedom were calculated using the Satterthwaite two-sample approximation.
