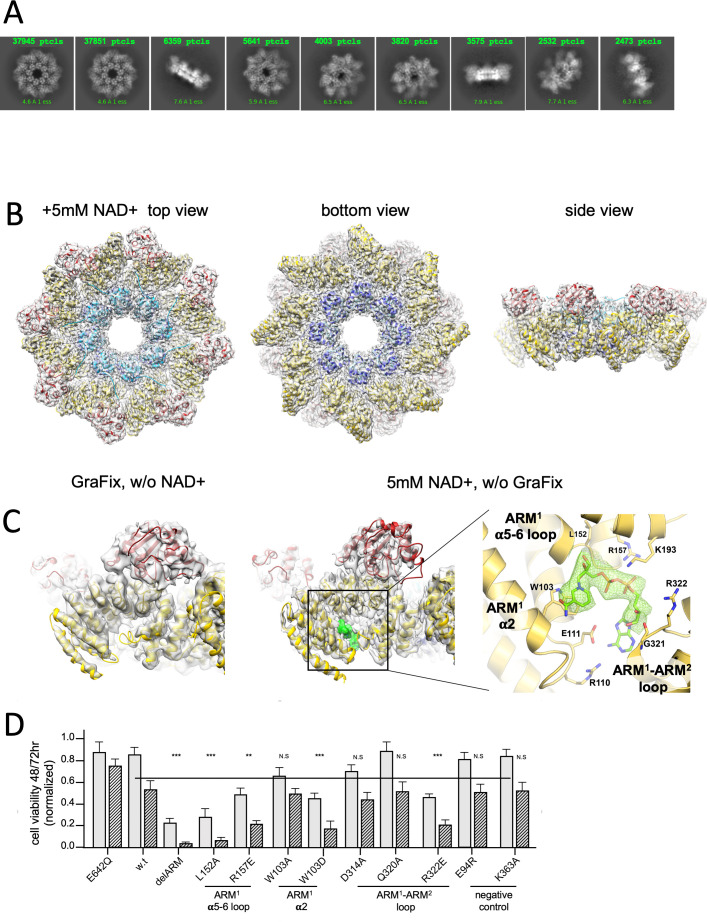
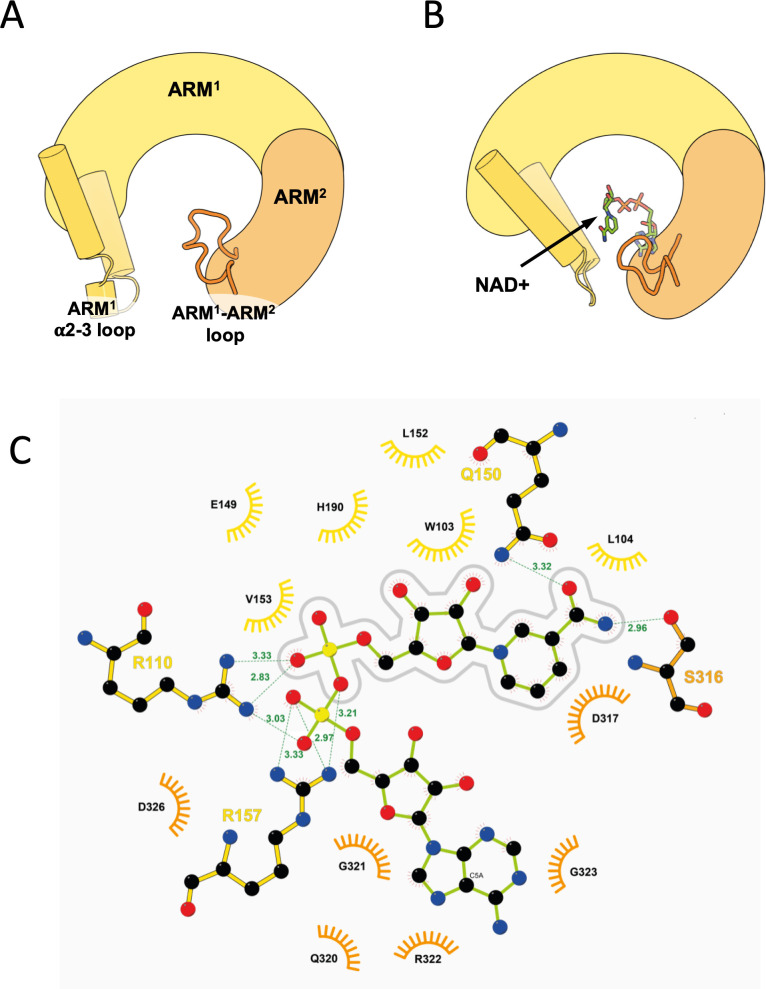
Figure 5. 3D structure reveals an inhibitory ARM allosteric NAD+ binding site.
(A) Selected representation of 2D class averages used for the 3D reconstruction of hSARM1E642Q supplemented with 5 mM of NAD+. The number of particles that were included in each class average is indicated. (B) Color coded (as in Figure 1A) protein model docked in a transparent 2.7 Å cryo-EM density map (gray). (C) When compared to the GraFix-ed map, without NAD+ supplement (left), an extra density appears at the ‘ARM horns’ region in the NAD+ supplemented map (middle, right). The extra density is rendered in green and an NAD+ molecule is fitted to it. The NAD+ is surrounded by three structural elements, as indicated on the right panel. The NAD+ directly interacts with the surrounding residues: the nicotinamide moiety is stacked onto the W103 side chain rings; the following ribose with L152 and H190; R110 forms salt bridge with phosphate alpha (proximal to the nicotinamide), while the beta phosphate (distal to the nicotinamide) forms salt bridges with R110 and R157. The map density at the distal ribose and adenosine moieties is less sharp, but clearly involves interactions with the ARM2 R322, G323, and D326. In this way, activation by nicotinamide mononucleotide, that lacks the distal phosphate, ribose, and adenosine, can be explained by binding to ARM1 while preventing the bridging interactions with ARM2. (D) Toxicity of the hSARM1 construct and mutants in HEK293F cells. The cells were transfected with hSARM1 expression vectors, as indicated. Viable cells were counted 48 (bars in gray) and 72 (bars in black stripes) hr post-transfection. Moderate reduction in cell viability due to ectopic expression of hSARM1w.t. becomes apparent 72 hr after transfection, when compared with the NADase attenuated hSARM1E642Q, while the ‘delARM’ construct marks a constitutive activity that brings about almost complete cell death after 3 days. Mutations at the ARM1 α5–α6 and ARM1–ARM2 loops induce cell death at a similar level as the ‘delARM’ construct, while the control mutations and W103A did not show increased activity (three biological repeats, Student’s t-test; ***p<0.001; *p<0.05; n.s: no significance).