Fig. 1. Experimental and analytical workflow from obtaining PBMCs to identifying potential immune cell markers.
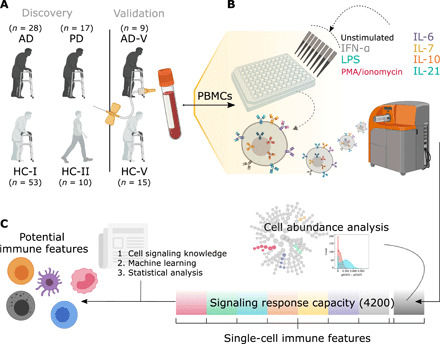
(A) In the discovery cohorts, whole blood was collected from 28 individuals with AD and 17 individuals with PD; AD was compared with the samples from 53 older HCs (HC-I), while PD samples were compared with a subset of those with age- and sex-matched HCs (HC-Isub). A different set of 10 younger HCs (HC-II) was included for examining age effects. In addition, an independent cohort of nine individuals with AD (AD-V) and 15 HCs (HC-V) was used for validation of the developed machine learning models without retraining. (B) PBMCs were either unstimulated or stimulated with IFN-α, IL-6, IL-7, IL-10, IL-21, LPS, or PMA/ionomycin. PBMCs were then bound with 21 metal-conjugated antibodies to surface markers and 15 metal-conjugated antibodies to intracellular signaling molecules before analysis by CyTOF. (C) Cell abundance was evaluated on PBMCs from an unstimulated condition. The stimulations and antibody probes generated a total of 4200 intracellular signaling responses (35 PMBC subtypes under eight stimulating conditions and assayed for 15 intracellular responses), which were used to identify the potential immune features with the aid of cell signaling knowledge, machine learning methods, and statistical analysis.
