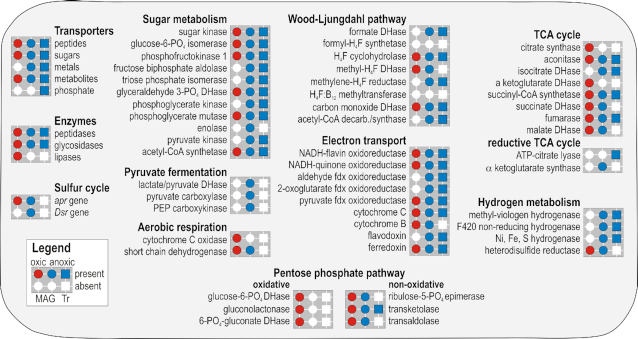
Figure 5.
Presence and absence of Chloroflexi assigned predicted proteins as detected in the present metagenomes and metatranscriptomes. Presence (colored) or absence (white) of metabolic functions (circles) and expressed genes (squares) identified in the metagenomes and metatranscriptomes at the oxic (red) and anoxic (blue) sites across all depths sampled. Presence/absence of substrate transporters, catabolytic enzymes, apr and Dsr genes are shown on the left hand side. Metabolic pathways listed correspond to sugar metabolism, pyruvate fermentation, aerobic respiration, Wood-Ljungdahl pathway, electron transport, tricarboxylic acid (TCA) cycle, hydrogen metabolism and oxidative and non-oxidative pentose phosphate pathway. The complete list of ORFs with assignment and coverage is available as Supplementary Data.