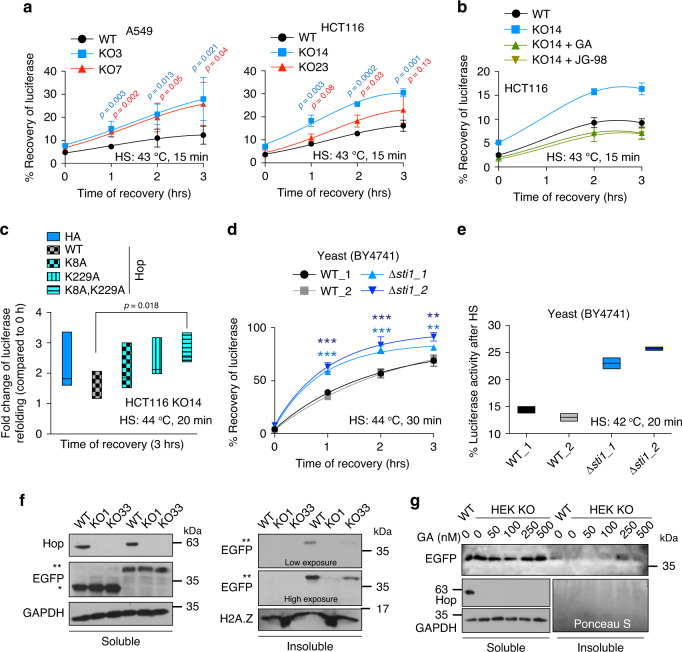
Fig. 6. Hsp70 and Hsp90 are functional in vivo even without Hop.
a In vivo refolding of heat-denatured luciferase. Luciferase activity before HS is set to 100%. Left panel, n = 4; right panel, n = 3 biologically independent samples. b In vivo luciferase refolding in HCT116 KO cells treated with inhibitors before and during the recovery phase (n = 3 biologically independent samples). c In vivo luciferase refolding in HCT116 KO cells exogenously overexpressing WT or TPR mutants of Hop. Non-HS controls for each sample are set 1-fold (n = 4 biologically independent samples). d In vivo refolding of heat-denatured luciferase in WT and Δsti1 yeast cells (BY4741 strain background; two different transformants each). ***p < 0.001 and **p < 0.01 statistically significant differences between WT and Δsti1 (n = 4 biologically independent samples). e Residual in vivo luciferase activity immediately after mild HS (n = 2 biologically independent samples). Luciferase activity before HS is set to 100%. f Solubility of aggregation-prone polyglutamine model protein. Immunoblots of Q23-EGFP (bands marked with *) and Q74-EGFP (**) from soluble and insoluble protein fractions. GAPDH and H2A.Z serve as loading controls for soluble and insoluble protein fractions, respectively. g Immunoblot of Q74-EGFP from soluble and insoluble protein fractions of KO cells treated with GA. The Ponceau S staining of the nitrocellulose filter serves as loading control for the insoluble protein fractions. For the line graphs, the data are represented as mean values ± SEM. For box plots, data are represented as the median values and edges of the box plots represent the range of the data. The statistical significance between the groups was analyzed by two-tail unpaired Student’s t-tests. Source data are provided as a Source Data file.