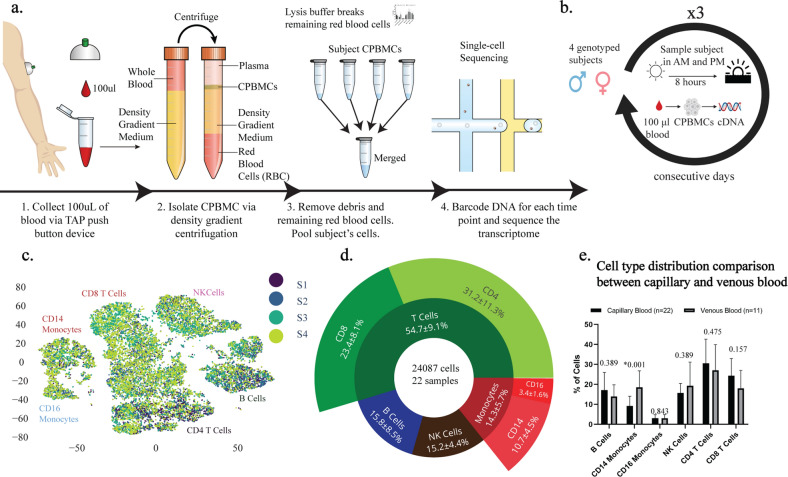
Figure 1.
Experimental workflow and consistency of capillary blood sampling. (a) Experimental workflow for capillary blood immune profiling. 1. Blood is collected using the TAP device from the deltoid. 2. Capillary peripheral blood mononuclear cells (CPBMCs) are separated via centrifugation. 3. Red blood cells are lysed and removed, and samples from different subjects are pooled together. 4. Cell transcriptomes are sequenced using single-cell sequencing. (b) Time-course study design. CPBMCs are collected and profiled from 4 subjects (2 male, 2 female) each morning (AM) and afternoon (PM) for 3 consecutive days. (c) 2-dimensional t-SNE projection of the transcriptomes of all cells in all samples. Cells appear to cluster by major cell type (Fig. S6) (d) Immune cell type percentages across all samples shows stable cell type abundances (includes cells without subject labels). (e) Cell type ratios between capillary blood from this study, and venous blood from 3 other studies were the same, with the exception of CD14+ Monocytes, which are more abundant in venous blood (FDR < 0.05, 2-sided student t-test, multiple comparison corrected) The q-values are displayed for each cell type comparison.