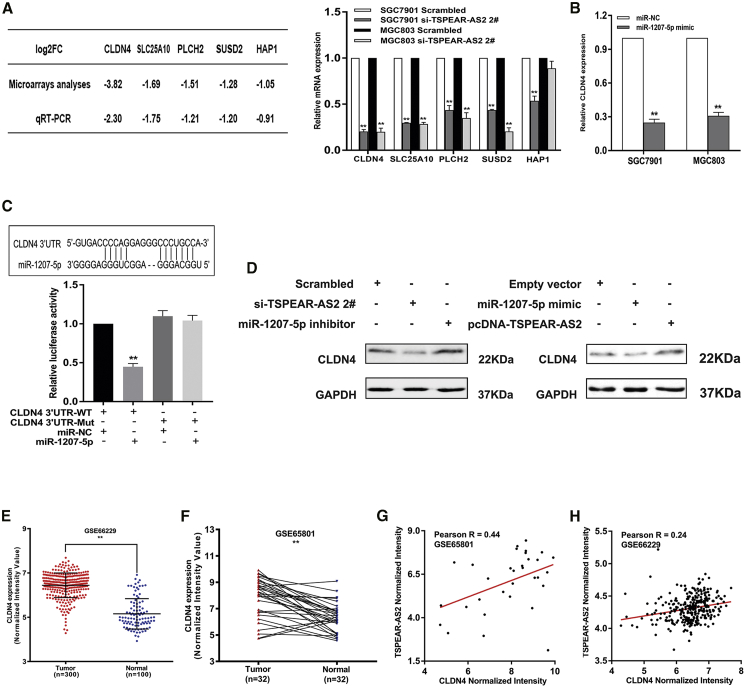
Figure 6.
lncRNA TSPEAR-AS2 Modulates CLDN4 Expression by Competing for miR-1207-5p in GC
(A) The qRT-PCR assays were conducted to validate the alteration of key genes in GC cells with TSPEAR-AS2 knockdown. (B) The effects of miR-1207-5p overexpression on CLDN4 expression in GC cells. (C) Luciferase activity in HEK293T cells cotransfected with miR-1207-5p or negative control and pmirGLO-CLDN4 3′UTR-WT or pmirGLO-CLDN4 3′UTR-Mut. (D) Western blot assay was used to detect CLDN4 protein level in GC cells upon miR-1207-5p mimic or pcDNA-TSPEAR-AS2 treatment (right), and in the absence of TSPEAR-AS2 expression or miR-1207-5p inhibitor (left). (E) The level of CLDN4 in gastric cancer tissues and normal tissues was measured in the data provided from GEO (GSE66229) datasets. (F) The level of CLDN4 in gastric cancer tissues and normal tissues was measured in the data provided from GEO (GSE65801) datasets. (G) The relationship between CLDN4 and TSPEAR-AS2 in GC tissues was determined based on the data from GSE65801 datasets. (H) The relationship between CLDN4 and TSPEAR-AS2 in GC tissues was determined based on the data from GSE66229 datasets.