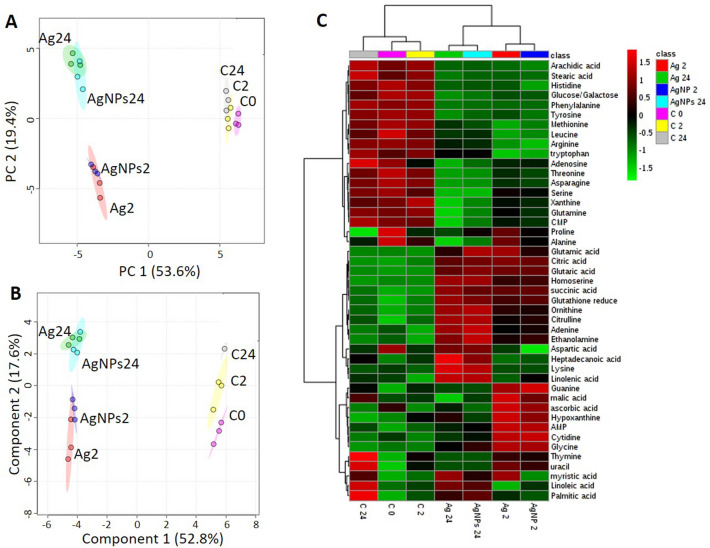
Figure 4.
(A) Principal component analysis (PCA) and (B) partial least-squares discriminate analysis (PLS-DA) score plots of metabolic profiles in P. malhamensis treated with AgNPs and dissolved Ag, and untreated controls. (C) Clustering metabolites and samples shown in a heat map (Euclidean distance and Ward clustering algorithm). Data were normalized by using probabilistic quotient normalization by untreated control group at time 0 (C0), log transformed and autoscaled. Exposure to 40.7 μg L−1AgNO3 for 2 h (Ag2) and 24 h (Ag24); exposure to 1 mgL−1AgNPs for 2 h (AgNPs2) and for 24 (AgNPs24); C2: unexposed controls at time 2 h; C24: unexposed controls at time 24 h. The score plots and heatmap are generated by MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/)72.