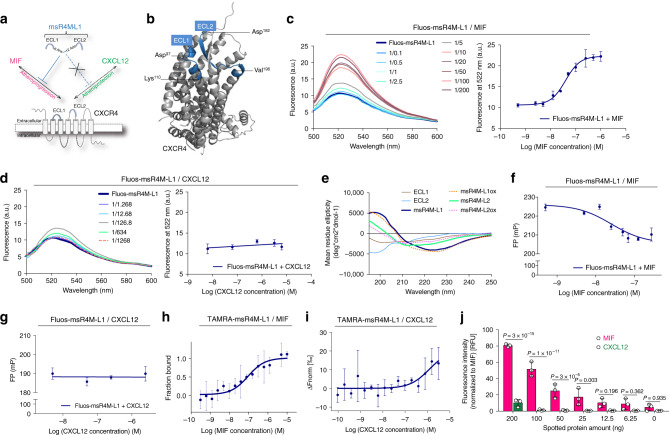
Fig. 1. The CXCR4 ectodomain mimic msR4M-L1 selectively binds to MIF but not CXCL12.
a Schematic summarizing the design strategy to utilize extracellular loop moieties of CXCR4 to engineer a soluble mimic that binds MIF but not CXCL12. b Ribbon structure of human CXCR4 based on the crystal structure according to PDB code 4RWS30. Sequences of extracellular loops ECL1 and ECL2 that were found to interact with MIF according to peptide array mapping25 are highlighted in blue, and the N- and C-terminal residues of the ECL1 and 2 peptides are indicated. c Nanomolar affinity binding of msR4M-L1 to MIF as determined by fluorescence spectroscopic titrations. Emission spectra of Fluos-msR4M-L1 alone (blue; 5 nM) and with increasing concentrations of MIF at indicated ratios are shown (left panel; representative titration); binding curve derived from the fluorescence emission at 522 nm (right panel). d msR4M-L1 does not bind to CXCL12. Same as c but titration performed with increasing concentrations of CXCL12. e Conformation of CXCR4 ectodomain mimics as determined by far-UV CD spectroscopy. Mean residue ellipticity plotted over the wavelength between 195 and 250 nm. f–g Nanomolar binding of msR4M-L1 to MIF (f) but not CXCL12 (g) as determined by fluorescence polarization (FP). The FP signal of 5 nM Fluos-msR4M-L1 (as mP) is plotted over varied ligand concentration as indicated. h, i Binding of msR4M-L1 to MIF (h) but not CXCL12 (i) as confirmed by microscale thermophoresis (MST). The fraction of chemokine bound or normalized fluorescence change (ΔFnorm) to 100 nM TAMRA-msR4M-L1 is plotted against increasing concentrations of MIF or CXCL12, respectively. j Binding analysis between TAMRA-msR4M-L1 and MIF versus CXCL12 as determined by dot blot titration. Quantification from three independent blots (see representative blot in Supplementary Fig. 8). Data in c–d (right panel) and f–h are reported as means ± SD from three independent experiments; data in i are means ± SD from five independent experiments. Statistical analysis (j) was performed with two-way ANOVA and Sidak correction. CXCR4, CXC motif chemokine receptor-4; msR4M-L1, MIF-specific CXCR4 mimic-L1; MIF, macrophage migration-inhibitory factor. Source data are provided as a Source Data file.