Highlights
-
•
LncRNA LINC00355 promotes colon cancer malignancy.
-
•
LncRNA LINC00355 positively regulates ITGA2 via recruiting GTF2B.
-
•
LncRNA LINC00355 positively regulates GTF2B-mediated ITGA2 to promote colon cancer.
-
•
This study proposes a novel targeted strategy for cancer treatment.
Keywords: Colon cancer, Long non-coding RNA00355, General transcription factor II-B, Integrin alpha-2, Invasion, Migration
Abstract
Long non-coding RNAs (LncRNAs) can regulate physiological and pathological functions, exhibiting a wide range of roles in cell biology. Moreover, many lncRNAs are dysregulated in various cancers, including colon cancer. In this study, we investigated the role of the lncRNA LINC00355 in colon cancer, after first establishing its interaction with GTF2B, and ITGA2 on the LncMap database. The predicted relationships between the lncRNA LINC00355, GTF2B, and ITGA2 were identified using luciferase reporter assay, RIP, and ChIP experiments. Western blot analysis and RT-qPCR were applied to determine expression pattern of lncRNA LINC00355 and ITGA2 in colon cancer cells. Additionally, EdU, TUNEL, Cell-adhesion and Transwell assay was used for the detection of the effects of this axis on proliferation, apoptosis, adhesion, chemotaxis and metastasis. LncRNA LINC00355 targeted IGFBP2 through the recruitment of GTF2B. LncRNA LINC00355 was highly expressed in colon cancer cells, and overexpression of lncRNA LINC00355 increased the expression of IGFBP2 and GTF2B, and thereby promoted the proliferation, chemotaxis, invasion, and migration in colon cancer. In summary, downregulation of lncRNA LINC00355 in colon cancer inhibited tumor growth in colon cancer through effects on the GTF2B/IGFBP2 axis.
Introduction
Colon cancer is one of the most frequently reported causes of cancer-related death, accounting for 6.1% of cancer incidence and 9.2% of cancer mortality in 2018 [1]. Colon cancer often develops from a local adenoma in the intestinal epithelium, developing to an aggressive malignant tumor that may metastasize to the liver [2]. Its incidence is increasing, with a predicted annual incidence of 2.2 million newly diagnosed cases by 2030, which shall place a huge burden on society [3]. There is evidence that intestinal microbiota alterations are linked with the development of colon cancer [4]. While treatable if detected early, there is a great risk for distant metastasis and poor outcome, in part due to the lack of reliable metastatic biomarkers [5]. Recently, much attention has been drawn to emerging predictive biomarkers, which may prove to be of vital importance for the timely treatment of colon cancer [6]. Long non-coding RNAs (lncRNAs) are among the emerging biomarkers predictive of cellular changes in the progression of colon cancer [7,8].
LncRNAs constitute a class of the allogeneic RNAs, which include intergenic lncRNAs, antisense transcripts, and enhancer RNAs [9]. Recent years have seen a growing appreciation of the importance of lncRNAs in the control of cell growth, apoptosis and metastasis [10], and an awareness that alterations in the function of lncRNAs contribute to tumor growth in many cancers [11]. Therefore, analysis of lncRNA alterations enables obtaining a better understanding of initiation and progression in colon cancer. For example, silencing of lncRNA LINC00659 is reported to inhibit cell growth and augment apoptosis in colon cancer [12]. Interestingly, lncRNA LINC00355 has poor expression in colon cancer, thus correlating inversely with disease progression [13], and presenting it as a noninvasive marker for colon malignancies. Moreover, the present study revealed integrin alpha-2 (ITGA2) to be a target of lncRNA LINC00355. Integrins are a family of heterodimeric cell surface adhesion molecules, each composed of one of eight possible beta-strands and one of 18 possible alpha-chains [14], which can potently mediate biological processes such as cell adhesion, migration, and proliferation [15]. In particular, ITGA2 contains a highly conserved plexin-brain signaling-integrin (PSI) domain [16]. Findings obtained from some studies have shown that ITGA2 has an extensive impact on apoptosis in a variety of cancers such as gastric cancer [17,18] and glioblastoma [19]. ITGA2 has been reported to play an important role in a variety of tumors, including the promotive effect on tumor proliferation and migration. For example, ITGA2 promotes tumor migration and invasion by ITGA2 in hepatocellular carcinoma via effects on the extracellular matrix, and the interaction of extracellular matrix is greatly reduced after blocking ITGA2 on the cell surface [17,20]. In our study, we attempted to determine new aspects of ITGA2 as a potential therapeutic target on colon cancer, focusing on interactions with transcription factors (TFs). For example, previous research reported that TBX5 is a novel tumor suppressor gene whose epigenetic inactivation is closely related to colon cancer [21]. Additionally, TF c-My and CDX2 play pivotal roles in regulation in colon cancer [22]. At the same time, general transcription factor II-B (GTF2B), a member of TF, has been identified as marker for the diagnosis of colon cancer [23]. Intriguingly, results from the lncMAP database implicate GTF2B in the regulation of lncRNA LINC00355 expression. Based on the above results, we aimed to test the hypothesis that the lncRNA LINC00355 may regulate ITGA2 via recruiting GTF2B to promote tumor growth in colon cancer.
Methods
Ethics statement
The institutional Experimental Animal Ethics Committee approved our study. All animal studies were undertaken in accordance with the principle to minimize the number and discomfort of the experimental animals.
Bioinformatics analysis to predict the differentially expressed genes (DEGs)
Clinical data and transcriptome data for colon cancer were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). DEGs along with lncRNA LINC00355 expression in colon cancer cells were identified by means of the limma package in R language, where |log2 fold change (FC)| > 2.0 and FDR-Val < 0.05 served as the threshold. LncRNALINC00355 was predicted using LncMap database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/index.jsp), and its relationship with TF and target genes was identified.
Cell treatment
Colonic epithelial cell line CCD841CON and colon cancer cell lines SW480, HT29, SW620, COLO205, HCT116 and T84 (American Type Culture Collection, Manassas, VA, USA) were cultured with 1640 Dulbecco's Modified Eagle Medium (DMEM) comprised of 10% fetal bovine serum and penicillin-streptomycin solution diluted to 1:1 to a final concentration of 100 U/mL at 37 °C in a 5% CO2 incubator. The cells were detached with 0.25% trypsin whereupon they were plated into a 6-well plate (3 × 105 cells/well), and then collected upon attaining about 70 - 80% confluence. The cell line with highest expression lncRNA LINC00355 detected by RT-qPCR assay was used in subsequent experiments.
Cell lines in logarithmic growth phase were added to a 6-well plate (4 × 105 cells/well). Two lentivirus vectors, including pLV-EGFP-N (overexpressing vector) and pSIH1-H1-copGFP (silencing of gene vector), were used in our study. When cells attained 30% confluence, we implemented infection with pSIH1-H1-copGFP expressing negative control (NC), pLV-EGFP-N,pLV-EGFP-N-LINC00355, pSIH1-H1-copGFP-LINC00355, pLV-EGFP-N-LINC00355 + pSIH1-H1-copGFP-short hairpin RNA (shRNA) targeting ITGA2, pSIH1-H1-copGFP-LINC00355 + pLV-EGFP-N-ITGA2, or pLV-EGFP-N-LINC00355 + pSIH1-H1-copGFP-sh-GTF2B. In the pre-experiment, we detected the silencing efficacy of three different shRNAs for ITGA2 and GTF2B to choose the shRNA with best silencing efficacy, and sh-ITGA2-1 and sh-GTF2B-1 were selected for the experiments (Supplementary Fig. 1).
RNA isolation and quantitation
Trizol (15596018, Beijing Solarbio Science and Technology Co., Ltd., Beijing, China) was used for total RNA extraction from liver tissues and cells. Primers used in the assay were prepared by TaKaRa Biotechnology Co., Ltd (Dalian, China) (Table 1). As per the instructions in the cDNA Synthesis Kit (K1622, Beijing Ya'anda Biotechnology Co., Ltd, Beijing, China), reverse transcription was implemented. The RT-qPCR assay was performed using a ViiA 7 PCR instrument (Daan Gene Co., Ltd., of Zhongshan, University Guangzhou, China). The relative expression of target genes was calculated with the 2−ΔΔCt method, with normalization to U6 snRNA and GAPDH mRNA levels, respectively.
Table 1.
Primer sequences for RT-qPCR.
| Gene | Primer sequence | |
|---|---|---|
| LINC00355 | F:GTTGGTGCCTGCTTTTCCAC | R:GGCGTGATACAACTGTCTGC |
| GAPDH | F:GCTCCCTCTTTCTTTGCAGC | R:ACCATGAGTCCTTCCACGAT |
Note: RT-qPCR, reverse transcription quantitative polymerase chain reaction; LncRNA LINC00355, long non-coding RNA00355; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; F, forward; R, reverse.
Fluorescence in situ hybridization (FISH)
After the cells had undergone culture in a 24-well plate (5 × 103 cells/well) for a period of 24 h, the supernatant was removed. The cells were fixed with methanol and added with PBS containing 0.5% Triton X-100. The pre-hybridization solution was blocked at 37 °C and hybridized with the lncRNA LINC00355 probe (M-030232-00) overnight at 37 °C. Next, hybridization solution was used for cell washing in the dark at 42 °C, followed by DAPI staining. The experimental specimens were observed under a laser confocal microscope (LEXT OLS4100, Olympus, Tokyo, Japan).
Western blot analysis
Total issue or cellular protein was isolated using RIPA lysis buffer. After separation by SDS-PAGE, the isolated proteins were transferred to PVDF membranes, and then blocked for 1 h with 5% BSA. The PVDF membrane and diluted primary antibodies to ITGA2 (ab133557, 1:50,000, Abcam, Cambridge, UK) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab8245’ 1:800, Abcam) were incubated overnight at 4 °C. After washing, the membrane underwent additional incubation with goat anti-rabbit IgG (ab205718, 1:20,000, Abcam, Cambridge, UK) labeled with horseradish peroxidase (HRP) for 1 h. Next, the membrane was developed by conventional methods and exposed, followed by gray scale analysis with ImageJ 1.48 u Software (National Institutes of Health). The relative ratio was calculated, with GAPDH serving as the internal reference.
Targeting relationship evaluated by dual luciferase reporter assay
The promoter sequence and the full sequence of ITGA2 was obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/gene).The promoter region of ITGA2 was cloned into pmirGLO (attained from Promega, Madison, WI, USA) luciferase vector, after which a recombinant vector of ITGA2-wild type (WT) (pmirGLO-ITGA2 prom WT) was obtained. Moreover, sh-GTF2B was obtained from Shanghai GenePharma Co., Ltd (Shanghai, Suzhou, China). pmirGLO-ITGA2 prom WT was co-transfected into cells with negative control (NC), sh-GTF2B + NC, pLV-EGFP-N-lncRNA LINC00355, GTF2B, pLV-EGFP-N-lncRNA LINC00355 + GTF2B, and sh-GTF2B + pLV-EGFP-N-lncRNA LINC00355, respectively. After transfection for 24 h, luciferase activity was measured using Renilla luciferase used as an internal reference using the dual luciferase reporter gene assay system (Promega, Madison, WI, USA).
ChIP assay
Cells were fixed with 4% formaldehyde and sonicated, followed by addition of rabbit anti-human GTF2B antibody (ab109106, 1:100, Abcam, Cambridge, UK) to combine with the GTF2B-ITGA2 promoter. Next, Protein Agarose/SaLmon Sperm DNA was added to precipitate the complex of GTF2B antibody and GTF2B-ITGA2 promoter. Precipitated DNA-protein complex was eluted and de-crosslinked, after which the DNA fragments were purified and analyzed by RT-qPCR.
RIP experiment
Cells were lysed in RIP lysis buff. Rabbit anti-human GTF2B antibody (ab109106, 1:100, Abcam, Cambridge, UK) was incubated with magnetic beads to the bead-GTF2B antibody complex, which was then mixed with the lysed cells. The magnetic bead-GTF2B antibody-GTF2B-LINC00355 complex was obtained by immunoprecipitation, and the immunoprecipitated RNA was isolated and purified, and then measured by RT-qPCR assay.
EdU assay
After seeding in 24-well plates, with 3 replicate wells set in each group, the cells were incubated with EdU at a final concentration of 10 μM, and then incubated for 2 h. Following fixation in PBS containing 4% paraformaldehyde for 15 min, the cells were incubated with 0.5% Triton-100 in PBS for 20 min, followed by 2 washes with PBS containing 3% BSA after the incubation. The washed cells were stained at room temperature in the dark for 30 min, followed by two washes in PBS containing 3% BSA, followed by DAPI staining. The positive cells in each field of view were counted under a FM-600 fluorescence microscope (Shanghai Pudan Optical Chemical Instrument Co., Ltd.).
TUNEL assay
Cells were seeded in 24-well plates. Upon attaining the logarithmic growth phase, cells were subject to transfection with the interfering sequence and cultured for 48 h. After supernatant removal, the cells were rinsed with PBS rising and fixed in 4% paraformaldehyde fixation for 15 min, followed by immersion in PBS. Cell apoptosis was assessed with the TUNEL Apoptosis Detection Kit (Boehringer Mannheim Gmbh, Mannheim, Germany) as per the manufacturer's instructions.
Cell-adhesion assay
The target tumor cells were incubated in a digestive solution (Sigma-Aldrich, St. Louis, MO, USA). Next, the cell suspension was centrifuged, whereupon the supernatant was removed, and the pellet was washed and resuspended with Hank's solution. Portions of cell suspension (100 μL, containing about 15,000 cells) was seeded in a 96-well flat-bottom non-cell culture plate pre-coated with rat tail collagen type I CoL (4 μg/well) overnight and blocked with 0.1% BSA. The next day, the non-adherent cells were removed by washing with Hanks' solution, and the number of adhered cells was counted by the BCA method.
Transwell assay
The cell invasion capacity was assessed with a Transwell chamber coated with Matrigel (MatrigeL: DMEM = 1:2), while migration was assessed in a Transwell chamber without Matrigel. The Transwell chamber was placed in an incubator at 37 °C for 4–5 h. After coagulation, the infected cells were diluted in 100 μL serum-free medium to prepare a cell suspension with a concentration of approximately 1 × 106 cells/mL. Then the cells were seeded in the apical chamber, with addition of 600 μL complete medium containing 20% FBS to the basolateral chamber. There were 3 duplicates for each well. After 24-h incubation at 37 °C with 5% CO2, the Transwell chamber was washed twice with PBS, fixed with 5% glutaraldehyde for 30 min, and stained with 0.1% gentian violet at 4 °C. The stained cells were counted in 5 randomly selected fields of view under an inverted fluorescence microscope (TE2000, Nikon, China).
Tumor xenografts in nude mice
Thirty-five 4–6-week old BALB/c nude mice (weighing 18–22 g, attained from Shanghai Lingchang Company, Shanghai, China) were reared in the animal housing of Guangdong Pharmaceutical University. Cells were infected with pSIH1-H1-copGFP-NC, pLV-EGFP-N, pLV-EGFP-N-LINC00355, pSIH1-H1-copGFP-LINC00355, pSIH1-H1-copGFP-sh-ITGA2, pSIH1-H1-copGFP-LINC00355 + pLV-EGFP-N-ITGA2 with titer of 2 × 108 PFU/mL, with a group without any treatment serving as a control. Mice were subcutaneously inoculated with 0.2 mL cell suspension containing 5 × 107 cells/mL under the left armpit with a syringe (1 mL). Tumor growth was observed and recorded on days 7, 14, 21, 28 and 35 after inoculation. Volume changes in nude mice were recorded using the formula: volume = π(a2b) /6, where a is the short diameter and b is the long diameter of the tumor. The terminal tumor mass was also weighed by a balance. After tumor formation, RNA and proteins were extracted from each tumor tissue of different groups for the subsequent RT-qPCR and Western blot analysis of LINC00355, GTF2B, and ITGA2 expression.
Statistical analysis
All data were expressed as mean ± standard deviation and analyzed by SPSS 19.0 statistical software (IBM Corp., Armonk, New York, USA). The difference between two groups was tested with paired t-test for paired data or unpaired t-test for unpaired data, after confirming normal distribution and homogeneity of the data. Differences between multiple groups were tested by one-way analysis of variance (ANOVA) with Tukey's post hoc test. Statistical analysis for data at different time points was conducted by repeated measure ANOVA, with Bonferroni corrections. For all statistical tests, a value of p < 0.05 was accepted as statistically significant.
Results
The potential participation of lncRNA LINC00355 in colon cancer progression
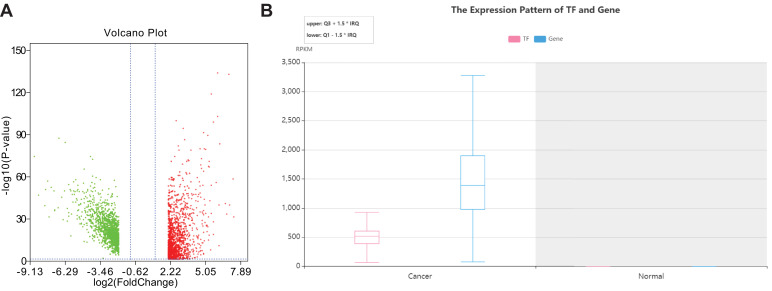
Clinical data and transcriptome data for colon cancer were downloaded from the TCGA database, which revealed 480 cases of cancer tissues and 41 cases of adjacent tissues. A total of 2400 DEGs were obtained, of which 1281 genes were highly expressed and 1119 were poorly expressed (Fig. 1A). The lncRNA LINC00355 is known to participate in the regulation of competing endogenous RNA (ceRNA) network, and shows very extensive alterations at each stage of colorectal cancer, implying it to be a promising target for colon cancer diagnosis and treatment [24]. LncRNA LINC00355 expression is associated with lymphatic metastasis, distant metastasis, and tumor stage, and lncRNA HULC was linked to tumor stage and lymphatic metastasis [13]. Present differential analysis results indicated that lncRNA LINC00355 was upregulated in colon cancer (Table 2).
Fig. 1.
LncRNA LINC00355 affects colon cancer via positively regulating ITGA2-mediated GTF2B. A, Volcano map representing differentially expressed lncRNAs related to colon cancer, the abscissa indicates logFC, the ordinate indicates log10 p value; each point in the Figure represents a gene, where red dots indicate up-regulated genes, and green dots indicate downregulated genes; B, Expression pattern of TF and genes, the ordinate indicating the readings per kilobase length from a given gene per million reads, and the abscissa indicating group information.
Table 2.
Expression of lncRNA LINC00355 in colon cancer tissue samples.
| LncRNA symbol | logFC | AveExpr | t | P value | Regulated |
|---|---|---|---|---|---|
| lncRNA LINC00355 | 3.291689 | −2.06986 | 2.700689 | 0.007662 | Up-regulated |
To further investigate the regulatory mechanism of lncRNA LINC00355 in colon cancer, we employed the LncMap database to predict the possible targets of lncRNA LINC00355, in conjunction with literature screening. Existing literature has shown that ITGA2 had a significant effect on apoptosis in gastric cancer and glioblastoma [17,19]. Therefore, we selected ITGA2 as an entry point to investigate the possible TFs whereby lncRNA LINC00355 might regulate ITGA2 expression. Through the prediction results from the lncMAP database, we identified 4 TFs involved in the regulation of lncRNA LINC00355, namely E2F1, GTF2B, RAD21, and SMAD2 (Table 3). Additionally, it has been confirmed previously that GTF2B is a significant factor in the diagnosis of colon cancer [23]. GTF2B and ITGA2 were highly expressed in the present cancer samples compared with normal samples, as predicted by LncMap (Fig. 1B). Therefore, we hypothesized that lncRNA LINC00355 might promote the proliferation of colon cancer via regulating ITGA2 by GTF2B.
Table 3.
Possible participation of lncRNA LINC00355, GTF2B and ITGA2 in colon cancer predicted by LncMap database.
| Cancer type | LncRNA symbol | TF ID | TF symbol | Gene symbol | Mediated pattern |
|---|---|---|---|---|---|
| COAD | LINC00355 | ENSG00000101412 | E2F1 | ITGA2 | - + |
| COAD | LINC00355 | ENSG00000137947 | GTF2B | ITGA2 | + + + |
| COAD | LINC00355 | ENSG00000164754 | RAD21 | ITGA2 | + - |
| COAD | LINC00355 | ENSG00000175387 | SMAD2 | ITGA2 | + - |
Note: COAD, colon adenocarcinoma; LncRNA, long non-coding RNA; TF, transcription factors; LncRNA LINC00320, long intergenic non-protein coding RNA 355; E2F1, E2 promoter binding factor 1; GTF2B, general transcription factor II-B; Smad2, Sma- and Mad-related protein 2; ITGA2, Integrin alpha-2.
LncRNA LINC00355 is amplified in colon cancer
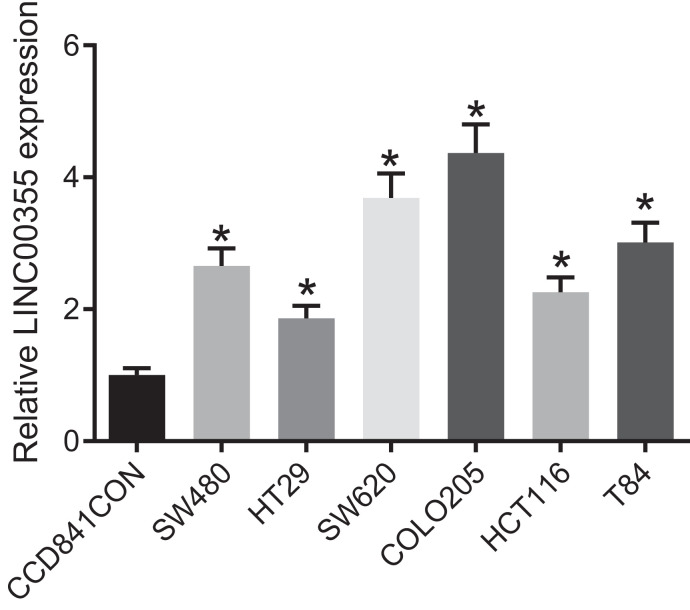
Colon cancer cell lines COLO205, SW620, T84, SW480, HCT116 and HT29 were selected to test the expression of lncRNA LINC00355. The result (Fig. 2) showed that all 6 colon cancer cell lines showed highly-expressed lncRNA LINC00355 relative to the normal colon epithelial cell line CCD841CON, with the highest seen in the COLO205 cell line. Thus, COLO205 was selected for further experiment.
Fig. 2.
LncRNA LINC00355 is overexpressed in colon cancer cells. Transcription level of lncRNA LINC00355 measured by RT-qPCR in colon cancer cell lines. *p < 0.05 vs. CCD841CON cell line. Data were expressed as mean ± standard deviation. Data comparison between groups was performed using one-way ANOVA with Tukey's post hoc test.
LncRNA LINC00355 promotes the colon cancer development
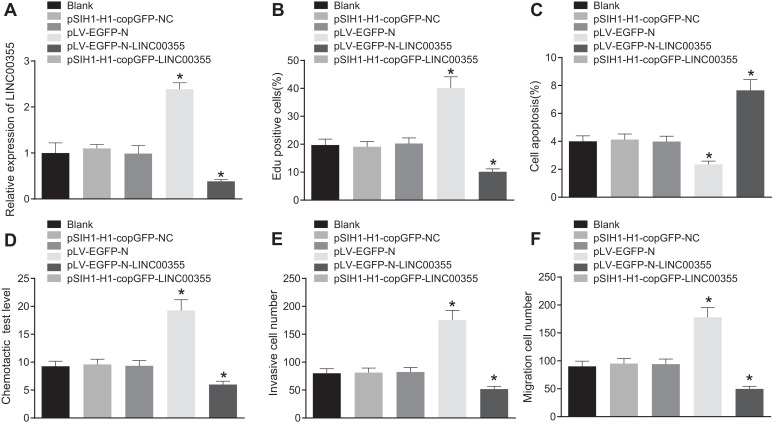
The results of RT-qPCR assay revealed that overexpression of lncRNA LINC00355 significantly increased lncRNA LINC00355 expression, while silencing lncRNA LINC00355 by the transfection of pSIH1-H1-copGFP-lncRNA LINC00355 significantly decreased lncRNA LINC00355 expression (p < 0.05; Fig. 3A). Subsequently, the EdU (Fig. 3B), TUNEL (Fig. 3C) and chemotaxis (Fig. 3D) experiments exhibited that knocking down lncRNA LINC00355 significantly inhibited cell proliferation, promoted apoptosis, and reduce cell chemotaxis, while overexpressing lncRNA LINC00355 significantly increased cell proliferation, inhibited apoptosis, and increased cell chemotaxis. Transwell assay results showed no significant difference in the number of invasive and migrating cells between the transfection of pSIH1-H1-copGFP-NC and pLV-EGFP-N compared with the blank group (p > 0.05). However, the number of invasive and migrating cells decreased significantly after treatment with pSIH1-H1-copGFP-lncRNA LINC00355. In contrast, the number of invasive and migrating cells treated with pLV-EGFP-N-LINC00355 increased significantly (Fig. 3E and F).
Fig. 3.
LncRNA LINC00355 contributes to the development of colon cancer. A, Transcription level of lncRNA LINC00355 measured by RT-qPCR in each tissue sample; B, EdU assay showing proliferation of the transfected cells; C, TUNEL assay for the detection of the apoptosis of the transfected cells; D, Absorbance in chemotaxis test. E, Number of invaded cells by Transwell. F, Number of migrated cells by Transwell. * p < 0.05 vs. cells treated without anything. Data were expressed as mean ± standard deviation. Data comparison between groups was performed using one-way ANOVA with Tukey's post hoc test.
LncRNA LINC00355 upregulates ITGA2 expression via recruiting GTF2B
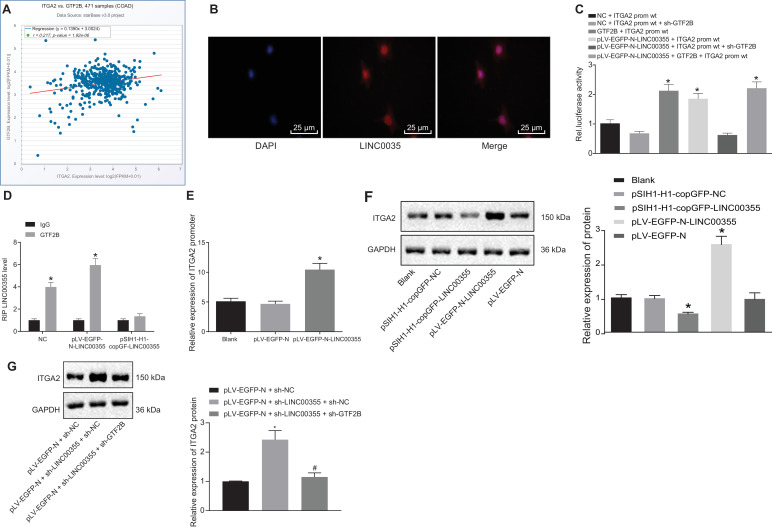
The 2 kb sequence upstream of the ITGA2 promoter was downloaded from the UCSC database, followed by prediction of the binding sites between ITGA2 promoter region and GTF2B using hTFtargetDatabase (http://bioinfo.life.hust.edu.cn/hTFtarget#!/), which yielded multiple binding sites in the promoter region (Supplementary Table 1). Further analysis of the correlation between GTF2B and ITGA2 expression in TCGA colon cancer samples revealed a significant positive correlation (Fig. 4A). The results of FISH analysis showed that lncRNA LINC00355 was present in the nucleus of malignant colon tumor cells (Fig. 4B). Analysis by dual luciferase reporter gene assay (Fig. 4C) showed that the luciferase activity in pLV-EGFP-N-LINC00355 + ITGA2 prom WT, GTF2B + ITGA2 prom WT and pLV-EGFP-N-LINC00355 + GTF2B + ITGA2 prom WT were all significantly increased compared with NC + ITGA2 prom WT (p < 0.05), yet no significant difference in luciferase activity was observed in pLV-EGFP-N-LINC00355 +ITGA2 PROM WT + sh-GTF2B compared with NC+ITGA2 PROM WT+ sh-GTF2B (p > 0.05). This result indicated that lncRNA LINC00355 played a regulatory role on ITGA2 through GTF2B.
Fig. 4.
LncRNA LINC00355 promotes expression of ITGA2 via recruiting GTF2B. A, Correlation analysis of GTF2B and ITGA2 in colon cancer. The abscissa represents ITGA2 expression while the ordinate represents GTF2B expression. Each point shows one sample. Correlation coefficient and correlation p value are listed above. B, Localization of lncRNA LINC00355 in cells by FISH; C, Luciferase signal of lncRNA LINC00355 in COLO205 cells detected by dual luciferase reporter gene assay; D, lncRNA LINC00355 expression in COLO205 cells detected by RIP test; E, relative expression of ITGA2 promoter determined by CHIP assay; F, Expression of ITGA2 in COLO205 cells measured by Western blot assay. G, ITGA2 protein expression detected by Western blot analysis. * p < 0.05 vs. cells infected with NC + ITGA2 prom WT, IgG, blank, or pLV-EGFP-N + sh-NC. # p < 0.05 vs. pLV-EGFP-N-LINC00355 + sh-NC. Data were expressed as mean ± standard deviation. Data were in compliance with normal distribution and homogeneity between two groups and were compared using an unpaired t-test. Data comparison between groups was performed using one-way ANOVA with Tukey's post hoc test.
The RIP test (Fig. 4D) displayed that lncRNA LINC00355 could directly bind to GTF2B protein, and ChIP assay results (Fig. 4E) showed that LINC00355 binding to GTF2B was enriched in the ITGA2 promoter region. Overexpression of lncRNA LINC00355 could increase the expression of ITGA2 protein as determined by Western blot assay (Fig. 4F and G). When LINC00355 was overexpressed while GTF2B was silenced simultaneously (Fig. 4H), the promoting effects of LINC00355 on ITGA2 were blocked. These results suggested that GTF2B/ITGA2 was the downstream axis of LINC00355.
Up-regulation of lncRNA LINC00355 enhances proliferation, migration, and invasion of colon malignant tumor cells via promoting GTF2B-mediated ITGA2
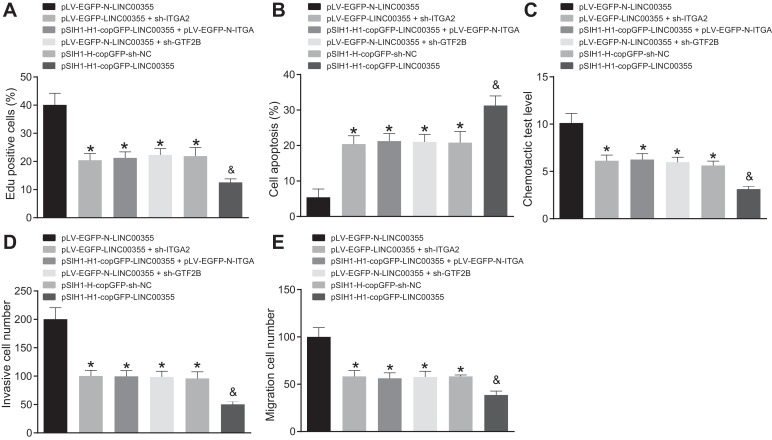
The results of EdU depicted the promoting effect of lncRNA LINC00355 overexpression in COLO205 cells on ITGA2 protein expression through GTF2B, with consequently enhanced cell proliferation, whereas deletion of lncRNA LINC00355 inhibited the expression of ITGA2 protein, and significantly reduced the proliferative rate (Fig. 5A). TUNEL assay results showed that knocking down lncRNA LINC00355 in COLO205 cells significantly inhibited ITGA2 expression and significantly increased the apoptosis rate, while overexpression of lncRNA LINC00355 provoked opposite effects (Fig. 5B).
Fig. 5.
Overexpressing the lncRNA LINC00355 enhances proliferation, invasion and migration of colon malignant tumor cells via promoting GTF2B-mediated ITGA2. A, Left: proliferation of the transfected cells detected by EdU assay; Right: proliferation of transfected cell were detected by EdU assay; B, Left: apoptosis of the transfected cells detected by TUNEL assay; Right: apoptosis of transfected cells detected by EdU assay; C, Chemotactic absorbance; D, the number of migrated cells in each transfected-cells measured by Transwell assay; E, the number of invaded cells in each transfected-cells measured by Transwell assay. * p < 0.05 vs. pLV-EGFP-N-LINC00355; and p < 0.05 vs. pSIH1-H-copGFP-sh-NC. Data were expressed as mean ± standard deviation. Data comparison between groups was performed using one-way ANOVA with Tukey's post hoc test.
The chemotactic results displayed that knocking down lncRNA LINC00355 in COLO205 cells significantly inhibited ITGA2 expression and reduced the chemotactic adhesion of cells, while the overexpression of lncRNA LINC00355 promoted ITGA2 expression through GTF2B and strengthened the chemotactic adhesion of cells (Fig. 5C).
The results of Transwell assay (Fig. 5D and E) indicated that, compared to cells treated with pLV-EGFP-N-LINC00355, number of invasion and migration cells was lower after treatment with pLV EGFP-LINC00355 + sh-ITGA2, pSIH1-H1-copGFP LINC00355+pLV EGFP-N-ITGA2, pLV EGFP-N-LINC00355 + sh-GTF2B (p < 0.05). Relative to treatment with the pSIH1-H1-copGFP-NC, the number of invasion and migration significantly decreased in cells treated withpSIH1-H1-copGFP LINC00355. No significant difference was evident in the number of invasive and migrating cells upon treatment with pLV-EGFP-N-LINC00355 + sh-ITGA2 or pSIH1-H1-copGFP-LINC00355 + pLV-EGFP-N-ITGA2 in comparison to treatment with pLV-EGFP-N-LINC00355 + sh-GTF2B or pSIH1-H-copGFP-sh-NC (p > 0.05). This suggested that overexpressing lncRNA LINC00355 promoted the expression of ITGA2 through GTF2B, thus increasing the invasion and migration potentials of cells. Conversely, knocking down lncRNA LINC00355 inhibited the expression of ITGA2, thus decreasing the invasion and migration of cells. Taken together, these results revealed that lncRNA LINC00355 enhanced the expression of ITGA2 through GTF2B, thereby promoting the proliferation, migration, and invasion associated with colon malignant tumor cells and inhibiting apoptosis.
Overexpression of lncRNA LINC00355 promotes tumor growth in nude mice
To further characterize the impact of lncRNA LINC00355, GTF2B and ITGA2 expression on the growth of colon malignant tumor in vivo, we analyzed the tumor volume and weight in nude mice that were injected with transfected cells. Results indicated no difference in tumor growth in mice injected with cells treated with pSIH1-H1-copGFP-NC, pLV-EGFP-N or pSIH1-H1-copGFP-LINC00355 + pLV-EGFP-NITGA2 relative to the blank treatment group. A trend towards slower tumor growth was noted in mice injected with cells treated with pSIH1-H1-copGFP-LINC00355 or pSIH1-H1-copGFP-sh-ITGA2 relative to that in the blank group, as well as smaller volume (seen in Fig. 6A) and mass (seen in Fig. 6B), while tumor growth was faster, attaining a larger terminal volume (seen in Fig. 6A) and mass (seen in Fig. 6B) in the mice injected with cells treated with pLV-EGFP-N-LINC00355 than that in blank group (All p < 0.05). Thus, overexpression of lncRNA LINC00355 accelerated tumor growth, and knockout of lncRNA LINC00355 or inhibition of ITGA2 inhibited tumor formation in vivo. Further RT-qPCR and Western blot (Fig. 6C and D) results showed no significant difference in the lncRNA LINC00355 expression and ITGA2 protein level in the tumor tissues of mice with pSIH1-H1-copGFP-NC and pLV-EGFP-N treatment compared with the mice used as blank control. The mice with pLV-EGFP-N-LINC00355 treatment showed the highest lncRNA LINC00355 expression and ITGA2 protein level, whereas lowest levels were seen in the mice with pSIH1-H1-copGFP-LINC00355 treatment. In the mice undergoing pSIH1-H1-copGFP-sh-ITGA2 treatment, the expression of LINC00355 exhibited no significant difference, while the protein level of ITGA2 was decreased. In the mice with pSIH1-H1-copGFP-LINC00355 + pLV-EGFP-N-ITGA2 treatment, the expression of LINC00355 was decreased, while the protein level of ITGA2 was not significantly altered.
Fig. 6.
Knocking down lncRNA LINC00355 inhibited tumor formation in nude mice. A, Growth volume and size of tumor in nude mice injected with transfected-cells; B, Mass of tumor in nude mice injected with transfected-cells. * p < 0.05 vs. the blank group. C, LINC00355 expression in tumor tissues detected by RT-qPCR. D, ITGA2 protein expression in tumor tissues examined by Western blot analysis. Statistical analysis in B was realized using ANOVA of repeated measurements with Tukey's post hoc test. Data comparison was performed using one-way ANOVA with Tukey's post hoc test. n = 5.
Discussion
Most colon cancer patients present with distant tumor metastases, which presents a great challenge for treatment [25]. Therefore, sensitive non-invasive biomarkers promoting disease staging, detection, and predictive treatment outcomes are urgently needed to provide better survival and establish optimal treatment options [26]. Recently, evidence has suggested that abnormally expressed lncRNAs can promote the occurrence of pathological events in the pathway for the development of colon cancer [27], [28]–29]. Thus, obtaining a better knowledge in relation to the mechanisms of lncRNAs may prove decisive to uncover more effective diagnostic and therapeutic means in colon cancer. In the present work, we investigated the potential role of the lncRNA LINC00355 in colon cancer.
Multiple lncRNAs extensively expressed in colon cancers, thereby promoting colon cancer progression. For example, lncRNA MALAT1, an upregulated lncRNA in colon cancer, facilitates tumor growth in colon cancer cells [30]. Additionally, the lncRNA TUG1 is highly expressed and works as an oncogenic gene in colon cancer due to its role in enhancing cell proliferation and migration in colon cancer cells [31]. Intriguingly, our present results also demonstrated that lncRNA LINC00355 expression was frequently amplified in colon cancer, and had the capacity to trigger proliferation, chemotaxis, invasion and migration of colon cancers. More importantly, the present study found that blocking lncRNA LINC00355 expression arrested the proliferation, invasion and migration of colon cancer cells and promoted apoptosis. Likewise, a previous study has validated that inhibition of the lncRNA CCEPR impedes invasion, proliferation, migration and cell cycle progression in colon cancer cells [32]. Similarly, suppression of the lncRNA AWPPH expression could suppress cell proliferation in colon cancer [33].
A few previous studies have found that high expression of ITGA2 plays a role of tumor-promoting properties in many cancers. For instance, ITGA2 expression showed an upward trend in metastatic lymph nodes and distant metastases, and its elevation predicted shorter overall survival in gastric cancer patients [18]. Similar results have been reported in our study that ITGA2 was overexpressed in colon cancer and exhibited a protective effect on proliferation, invasion and metastasis of colon cancer cells. Furthermore, a previous study has identified high expression of ITGA2 in gastric cancer, and that blocking ITGA2 inhibits actin organization and cell migration in gastric cancer cells [17]. Consistent with these results, we also found that knockdown of ITGA2 contributed to inhibition of tumor growth in colon cancer. Intriguingly, the lncRNA ENST00000455974 exerts carcinogenic effects by up-regulating JAG2 in colon cancer [34], which was also partially consistent with our findings that lncRNA LINC00355 could positively regulate ITGA2 to promote colon cancer. These findings strongly suggested the potential role of lncRNA LINC00355and ITGA2 in tumorigenesis. However, their detailed regulatory mechanisms in colon cancer are not well characterized.
Previously conducted studies have suggested many tumor-associated antigens (TAA) such as 90 K [35], 4D10 [36] and GTF2B [23] may serve as targets for colon cancer therapy [37]. Moreover, the relationship between GTF2B, lncRNA LINC00355 and ITGA2 was verified in our study, which showed that overexpression of lncRNA LINC00355 was conducive to promoting the binding of GTF2B to ITGA2. After binding to GTF2B, lncRNA LINC00355 was enriched in the ITGA2 promoter region, thereby promoting its transcription and expression. All these findings collectively suggested that lncRNA LINC00355 could positively regulate ITGA2 via recruiting GTF2B. Besides, our data showed that knockdown of lncRNA LINC00355 suppressed the expression of ITGA2 that is positively regulated by GTF2B, thereby inhibiting the invasion, proliferation, and migration of colon cancer cells and promoting apoptosis.
Conclusion
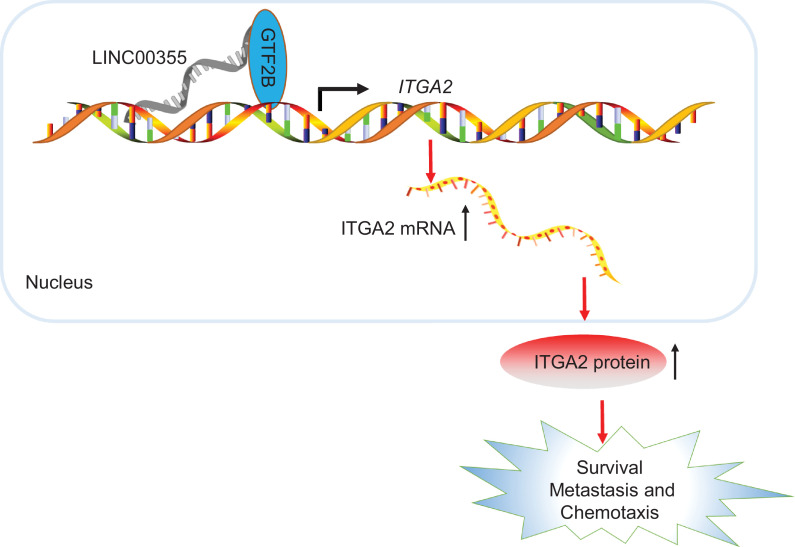
The present study determined the mechanistic roles of the lncRNA LINC00355/ITGA2/GTF2B axis in the regulation of colon cancer. In addition, we identified the effect of lncRNA LINC00355 in the occurrence and development of colon cancer in a nude mouse model. LINC00355 promoted cell invasion, proliferation, and migration in colon cancer by positively regulating ITGA2 through the recruitment of GTF2B (Fig. 7). In contrast, silencing of lncRNA LINC00355 impeded cell proliferation, chemotactic and metastasis. These results offer a potential basis for further research towards novel targeted therapy of colon cancer, with the caveat that our sample size was relatively small. Besides, it remains to be established how ITGA2 controls cell progression in colon cancer.
Fig. 7.
The lncRNA LINC00355 regulates ITGA2 via recruiting GTF2B thereby promoting the proliferation, invasion and metastasis in colon cancer. In colon cancer, high expression of lncRNA LINC00355 enhanced the expression of ITGA2 by recruiting GTF2B, thus promoting the survival, chemotaxis and metastasis of colon malignant tumors.
Supplementary Figure 1 The silencing efficacy of different shRNAs as detected by RT-qPCR. A, the silencing efficacy of three shRNAs for ITGA2; B, the silencing efficacy of three shRNAs for GTF2B.
CRediT authorship contribution statement
Zhiyan Ruan: Conceptualization, Visualization, Writing - review & editing. Hongling Deng: Conceptualization, Visualization, Writing - review & editing. Minhua Liang: Data curation, Validation, Writing - review & editing. Zhe Xu: Data curation, Validation, Writing - review & editing. Manxiang Lai: Writing - original draft, Writing - review & editing. Hong Ren: Writing - original draft, Writing - review & editing. Xiangliang Deng: Writing - review & editing. Xinguo Su: Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
Acknowledgments
We are willing to give our sincere gratitude to the reviewers owing to their helpful comments on this article.
Funding
This study was supported by the Foundation of Medical Science and Technology of Guangdong Province (A2019549 and A2020412); Innovation Team Project of Universities in Guangdong Province (2018GKCXTD005); Innovation Project of Universities in Guangdong Province (2019GKTSCX046); Scientific Project of Guangdong Food and Drug Vocational College (2019ZR06).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100947.
Contributor Information
Xiangliang Deng, Email: dxl@gdpu.edu.cn.
Xinguo Su, Email: suxg@gdyzy.edu.cn.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Varnat F., Duquet A., Malerba M. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol. Med. 2009;1:338–351. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold M., Sierra M.S., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 4.Kwong T.N.Y., Wang X., Nakatsu G. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018;155:383–390. doi: 10.1053/j.gastro.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 5.Hur K., Toiyama Y., Okugawa Y., Ide S., Imaoka H., Boland C.R., Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66(4):654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dienstmann R., Salazar R., Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J. Clin. Oncol. 2015;33(16):1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 7.White N.M., M.aher C.A. The potential use of lncRNAs found in the 8q24 region as biomarkers for colon cancer. Ann. Oncol. 2017;28:1688–1689. doi: 10.1093/annonc/mdx337. [DOI] [PubMed] [Google Scholar]
- 8.Liang Z.X., Liu H.S., Wang F.W., Xiong L., Zhou C., Hu T., He X.W., Wu X.J., Xie D., Wu X.R. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019;10(11):829. doi: 10.1038/s41419-019-2077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boon R.A., Jae N., Holdt L., Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets. J. Am. Coll. Cardiol. 2016;67(10):1214–1226. doi: 10.1016/j.jacc.2015.12.051. [DOI] [PubMed] [Google Scholar]
- 10.Martens-Uzunova E.S., Bottcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65(6):1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Yan X., Hu Z., Feng Y., Hu X., Yuan J., Zhao S.D., Zhang Y., Yang L., Shan W., He Q. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28(4):529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai K.W., L.o Y.H., Liu H. Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer. Mol. Cancer. 2018;17:72. doi: 10.1186/s12943-018-0821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu R., Shen X., Si Y., Fu Y., Zhu W., Xiao T., Fu Z., Zhang P., Cheng J., Jiang H. MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment. Aging Cell. 2018;17(4):e12794. doi: 10.1111/acel.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Franceschi N., Arjonen A., Elkhatib N. Selective integrin endocytosis is driven by interactions between the integrin alpha-chain and AP2. Nat. Struct. Mol. Biol. 2016;23:172–179. doi: 10.1038/nsmb.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong H., Shen B., Flevaris P., Chow C., Lam S.C., Voyno-Yasenetskaya T.A., Kozasa T., Du X. G protein subunit Galpha13 binds to integrin alphaIIbbeta3 and mediates integrin "outside-in" signaling. Science. 2010;327(5963):340–343. doi: 10.1126/science.1174779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu G., Zhang Q., R.eddy E.C. The integrin PSI domain has an endogenous thiol isomerase function and is a novel target for antiplatelet therapy. Blood. 2017;129:1840–1854. doi: 10.1182/blood-2016-07-729400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang Y.C., Wu H.Y., Lin Y.L., Tzou S.C., Chuang C.H., Jian T.Y., Chen P.R., Chang Y.C., Lin C.H., Huang T.H. Blockade of ITGA2 induces apoptosis and inhibits cell migration in gastric cancer. Biol. Proced. Online. 2018;20:10. doi: 10.1186/s12575-018-0073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong J., Wang R., Ren G., Li X., Wang J., Sun Y., Liang J., Nie Y., Wu K., Feng B. HMGA2-FOXL2 axis regulates metastases and epithelial-to-mesenchymal transition of chemoresistant gastric cancer. Clin. Cancer Res. 2017;23(13):3461–3473. doi: 10.1158/1078-0432.CCR-16-2180. [DOI] [PubMed] [Google Scholar]
- 19.Guo P., Moses-Gardner A., Huang J. ITGA2 as a potential nanotherapeutic target for glioblastoma. Sci. Rep. 2019;9:6195. doi: 10.1038/s41598-019-42643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C., Zeisberg M., Lively J.C., Nyberg P., Afdhal N., Kalluri R. Integrin alpha1beta1 and alpha2beta1 are the key regulators of hepatocarcinoma cell invasion across the fibrotic matrix microenvironment. Cancer Res. 2003;63(23):8312–8317. [PubMed] [Google Scholar]
- 21.Yu J., Ma X., C.heung K.F. Epigenetic inactivation of T-box transcription factor 5, a novel tumor suppressor gene, is associated with colon cancer. Oncogene. 2010;29:6464–6474. doi: 10.1038/onc.2010.370. [DOI] [PubMed] [Google Scholar]
- 22.Sakuma K., Aoki M., Kannagi R. Transcription factors c-Myc and CDX2 mediate E-selectin ligand expression in colon cancer cells undergoing EGF/bFGF-induced epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA. 2012;109(20):7776–7781. doi: 10.1073/pnas.1111135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villar-Vazquez R., Padilla G., Fernandez-Acenero M.J., Suarez A., Fuente E., Pastor C., Calero M., Barderas R., Casal J.I. Development of a novel multiplex beads-based assay for autoantibody detection for colorectal cancer diagnosis. Proteomics. 2016;16(8):1280–1290. doi: 10.1002/pmic.201500413. [DOI] [PubMed] [Google Scholar]
- 24.Tian S., Liu W., Pan Y. Long non-coding RNA Linc00320 inhibits glioma cell proliferation through restraining Wnt/beta-catenin signaling. Biochem. Biophys. Res. Commun. 2019;508:458–464. doi: 10.1016/j.bbrc.2018.11.101. [DOI] [PubMed] [Google Scholar]
- 25.Riihimaki M., Hemminki A., Sundquist J., Hemminki K. Patterns of metastasis in colon and rectal cancer. Sci. Rep. 2016;6:29765. doi: 10.1038/srep29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H., Zhang L., C.ogdell D.E. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F., Li Q., Wu X. Construction and analysis for differentially expressed long non-coding RNAs and microRNAs mediated competing endogenous RNA network in colon cancer. PLoS One. 2018;13(2) doi: 10.1371/journal.pone.0192494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X., Mi L., Dong J. Long intergenic non-protein-coding RNA 1567 (LINC01567) acts as a "sponge" against microRNA-93 in regulating the proliferation and tumorigenesis of human colon cancer stem cells. BMC Cancer. 2017;17:716. doi: 10.1186/s12885-017-3731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R., Li J., Yan X., Jin K., Li W., Liu X., Zhao J., Shang W., Liu Y. Long noncoding RNA plasmacytoma variant translocation 1 (PVT1) promotes colon cancer progression via endogenous sponging miR-26b. Med. Sci. Monit. 2018;24:8685–8692. doi: 10.12659/MSM.910955. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Wu Q., Meng W.Y., Jie Y., Zhao H. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J. Cell. Physiol. 2018;233(9):6750–6757. doi: 10.1002/jcp.26383. [DOI] [PubMed] [Google Scholar]
- 31.Liang Y., Rong X., Luo Y., Li P., Han Q., Wei L., Wang E. A novel long non-coding RNA LINC00355 promotes proliferation of lung adenocarcinoma cells by down-regulating miR-195 and up-regulating the expression of CCNE1. Cell Signal. 2020;66 doi: 10.1016/j.cellsig.2019.109462. [DOI] [PubMed] [Google Scholar]
- 32.Duan Y., Fang Z., Shi Z., Zhang L. Knockdown of lncRNA CCEPR suppresses colorectal cancer progression. Exp. Ther. Med. 2019;18(5):3534–3542. doi: 10.3892/etm.2019.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai J., Xu J., Zhao J., Zhang R. Downregulation of lncRNA AWPPH inhibits colon cancer cell proliferation by downregulating GLUT-1. Oncol. Lett. 2019;18(2):2007–2012. doi: 10.3892/ol.2019.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lao Y., Li Q., Li N. Long noncoding RNA ENST00000455974 plays an oncogenic role through up-regulating JAG2 in human DNA mismatch repair-proficient colon cancer. Biochem. Biophys. Res. Commun. 2019;508:339–347. doi: 10.1016/j.bbrc.2018.11.088. [DOI] [PubMed] [Google Scholar]
- 35.Hee Lee J., P.ark M.S., H.wang J.E. Dendritic cell-based immunotherapy for colon cancer using an HLA-A*0201-restricted cytotoxic T-lymphocyte epitope from tumor-associated antigen 90K. Cell Mol. Immunol. 2013;10:275–282. doi: 10.1038/cmi.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X.Y., Yao X.L., Song Y., Liu X.Y., Zhang P., Xu Z.S., Song L.H. Preparation and characterization of human colon tumor-associated antigen and its clinical significance. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2007;23(4):335–337. [PubMed] [Google Scholar]
- 37.Ploper D., De Robertis E.M. The MITF family of transcription factors: role in endolysosomal biogenesis, Wnt signaling, and oncogenesis. Pharmacol. Res. 2015;99:36–43. doi: 10.1016/j.phrs.2015.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.