Fig. 5.
Protein-based cobalt sensors.
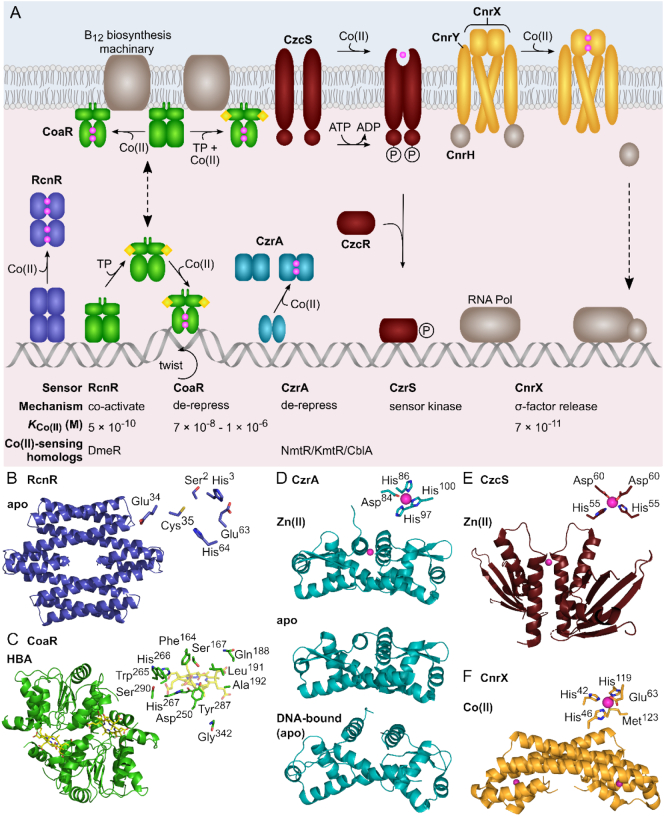
A, RcnR represses expression of rcnRAB (in E. coli and Salmonella, two RcnR tetramers bind the target site). RcnR can bind four Co(II) ions per tetramer, which weakens the affinity for DNA, de-repressing expression [83]. CoaR binds the coaT promoter in the absence of effector. Co(II) binding is predicted to induce a conformational change which distorts the operator-promoter, enabling recruitment of RNA polymerase and activation of expression. CoaR also harbours a tetrapyrrole (TP) binding domain including a hydrophobic patch which is capable of interacting with membranes [96]. Tetrapyrrole binding might tighten the affinity of CoaR for Co(II), and/or association with membrane-associated B12 biosynthetic machinery could confer a kinetic advantage for Co(II) and/or tetrapyrrole acquisition. CzrA represses expression of czrAB, and Co(II) binding weakens the DNA affinity of the CzrA dimer, alleviating repression. The structures of apo- and Zn(II)-CzrA are similar in the absence of DNA and an entropic mechanism for allosteric regulation has been demonstrated [103]. CzcS sensor histidine kinase detects periplasmic Co(II), and metal-binding induces autophosphorylation at a conserved histidine by Mg(II)-GTP. Transphosphorylation of the response regulator CzcR induces a conformational change promoting DNA-binding and activation of czcCBA [113]. CnrX is a transmembrane protein which forms a complex with CnrY and cytoplasmic CnrH. Co(II) binding to the periplasmic domain of CnrX, induces a conformational change which is transduced through the complex, releasing the CnrH extracytoplasmic function sigma factor. CnrH is recruited by RNA polymerase enabling expression of cnrCBA [112,113]. B–F, structural models of Co(II)-sensing proteins. B, Structural representation of RcnR tetramer (dimer of dimers) generated from PDB 5LYC. The ligands identified for co-ordination of Co(II) are shown. C, Dimeric representation of the deduced tetrapyrrole binding domain of CoaR bound to hydrogenobyrinic acid (HBA), modelled on the structure of CobH precorrin isomerase (PDB 1I1H) as described in [96] (main chain of Gly342 shown). The CoaR ligands which spatially overlay with the HBA-binding site of CobH are shown. D, The structures of Zn(II)- (PDB 2M30) and apo-CzrA (PDB 1R1U) reveal similar ‘open’ conformations which contrasts with the ‘closed’ conformation of the DNA-bound form (PDB 2KJB) [103]. The Zn(II) (and Co(II)) binding site at the dimer interface is shown. E, The sensor domain of P. aeruginosa Zn(II)-CzcS (PDB 5GPO) suggests Zn(II) binds to reciprocal residues at the dimer interface. His55, but not Asp60, is conserved in the Co(II)-responsive CzcS homolog from C. metallidurans [114]. F, The soluble periplasmic domain of CnrX (PDB 2Y3B) displays a similar structural fold to MerR-family sensors such as RcnR. The Co(II)-binding site is shown: a sulphur donor ligand is provided by a conserved methionine in contrast to the invariant cysteine in RcnR [83,120].