Fig. 4.
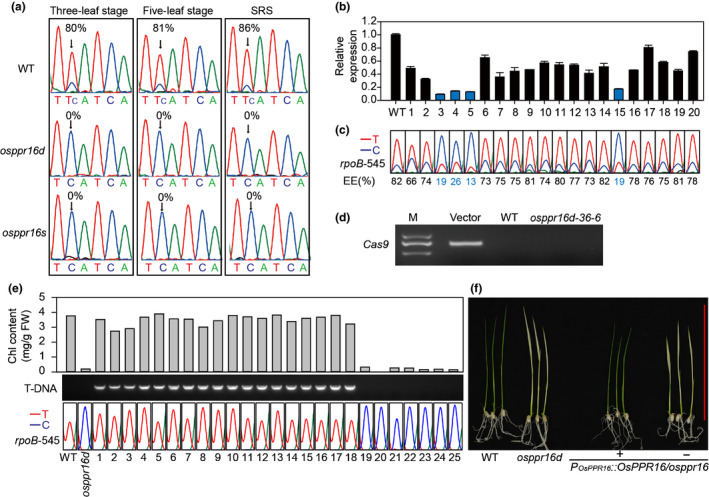
Editing analysis of rpoB‐545 in rice (Oryza sativa) wild‐type (WT), osppr16, RNA interference (RNAi), and complemented plants. (a) Editing efficiency of rpoB‐545 in WT and osppr16 leaves at three‐leaf, five‐leaf, and seed ripening stages. SRS, seed ripening stage. The arrows refer to the editing site. (b) Relative expression of OsPPR16 in RNAi transgenic plants, as determined by quantitative real‐time PCR. OsTPI was used as reference gene. (c) Editing efficiency of rpoB‐545 in OsPPR16 RNAi lines. EE, editing efficiency. (d) Assay for the Cas9 gene by PCR in osppr16d T1 plants. M, marker. Vector, pYLCRISPR‐osppr16d. The osppr16d‐36‐6 plant was confirmed as Cas9‐free line, and subsequently used to conduct complementation analysis. (e) Analysis of Chl content, transfer DNA (T‐DNA) presence, and RNA editing efficiency in WT, osppr16d, and T1 seedlings from a T0 complemented line at the three‐leaf stage. (f) Phenotypes of WT, osppr16d, and complemented T1 seedlings (POsPPR16::OsPPR16/osppr16). +, positive T1 seedlings segregated from a T0 complementation line; −, negative T1 seedlings segregated from a T0 complementation line. Bar, 10 cm.
