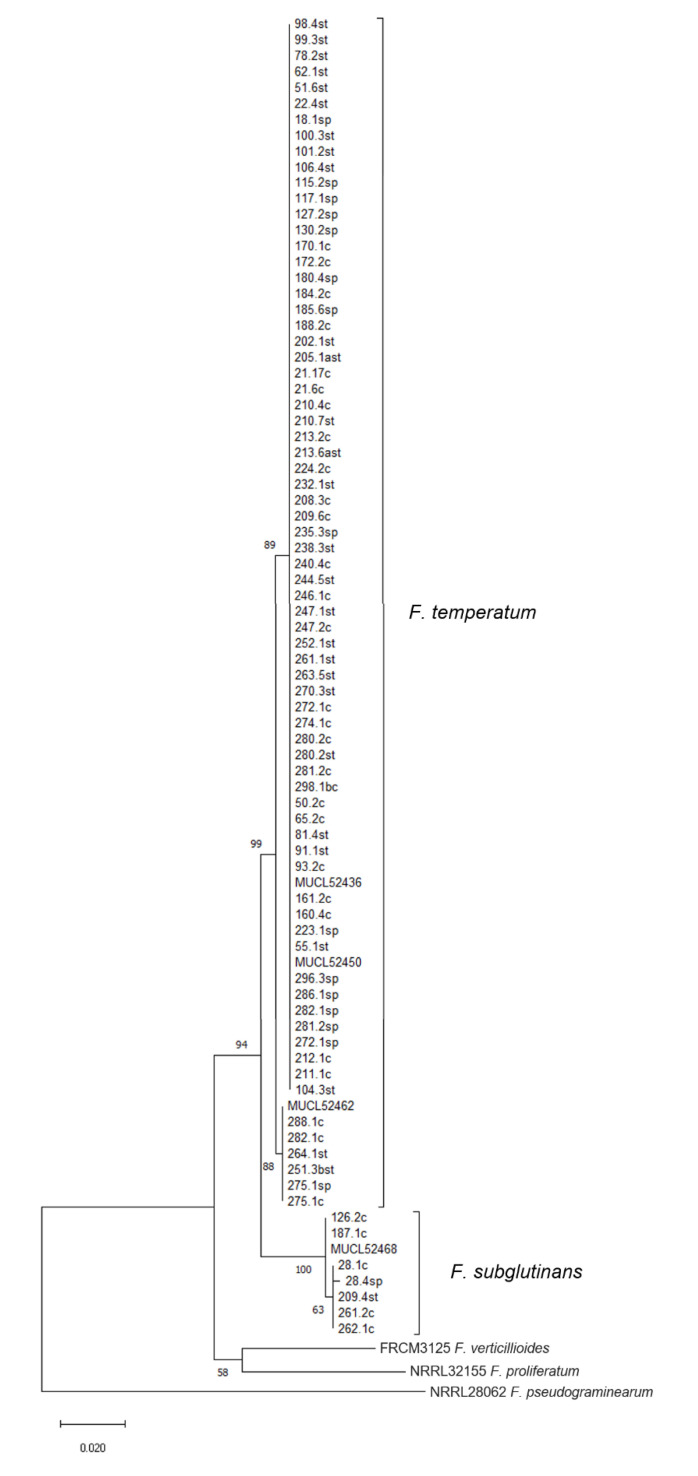
Figure 2.
Molecular phylogenetic analysis of translation elongation factor 1 alpha (TEF-1α) by the maximum likelihood method (1000 bootstrap replicates) [27]. Analysis was performed with ClustalW [25] in MEGA version 7.0.26 [26] with partial TEF-1α sequences of 72 isolates of F. temperatum and 7 isolates of F. subglutinans (Table S3), and references for F. temperatum MUCL52436, MUCL52450, MUCL52462, and F. subglutinans MUCL52468 [10]. We added additional references for F. proliferatum NRRL32155, F. verticillioides FRCM3125, and F. pseudograminearum NRRL26062 to scale phylogenetic separation. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Bootstrap values are presented next to the nodes. Nucleotide sequences have been subjected to Genbank, the accession numbers are presented in Tables S1 and S5.