Fig. 3 –
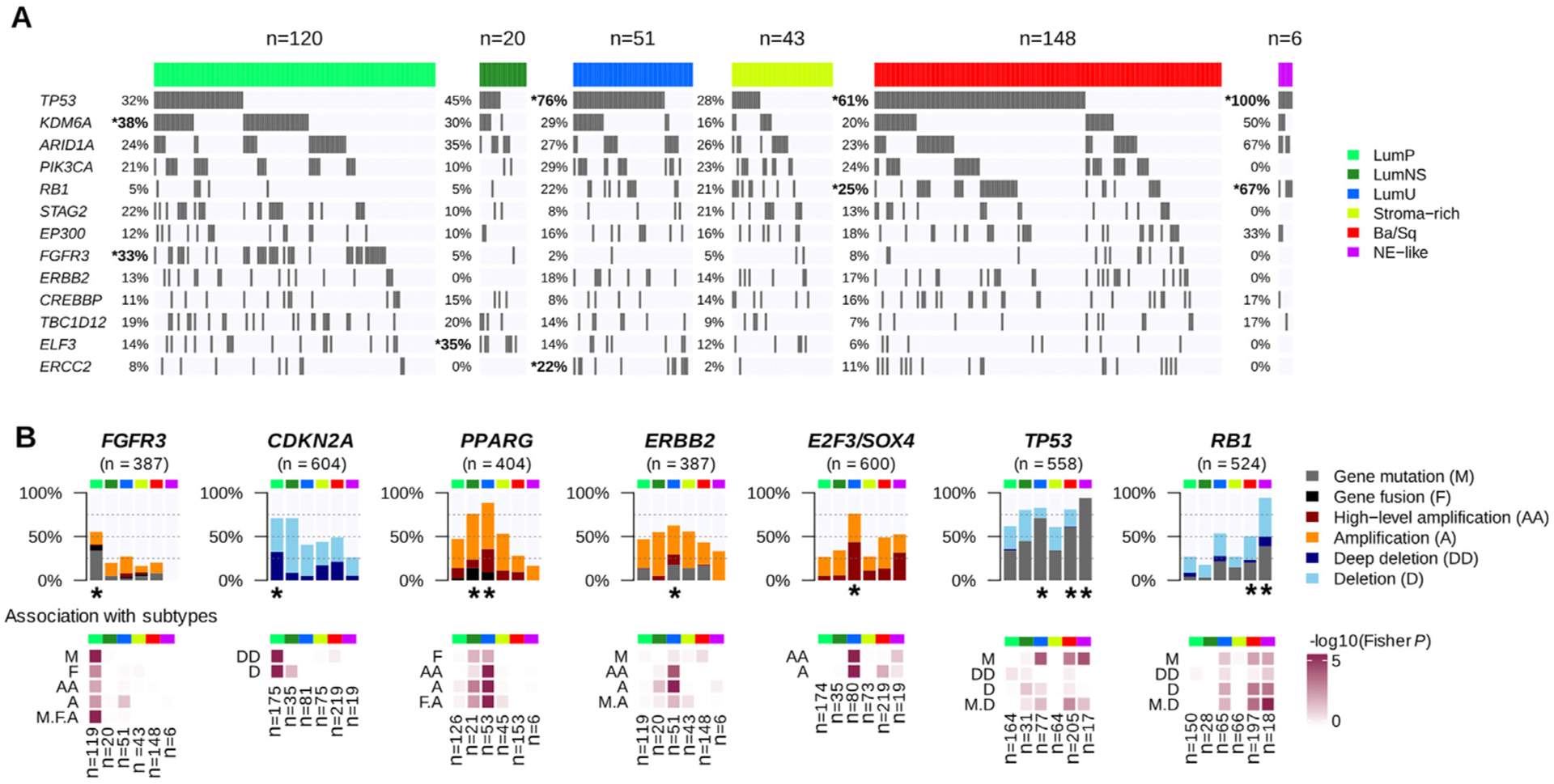
Genomic alterations associated with consensus classes. (A) We used the available exome data from 388 TCGA MIBC samples to study the association between consensus classes and specific gene mutations (see Supplementary Table 4 and Supplementary Fig. 3). The panel displays the 13 genes with MutSig q values <0.02 found in >10% of all tumors. Gene mutations that were significantly enriched in one consensus class are marked by an asterisk. (B) Combined genomic alterations associated with seven bladder cancer-associated genes and statistical association with consensus classes. Upper panels: main alteration types after aggregating CNA profiles (see Supplementary Table 5) from CIT (n = 87), Iyer (n = 58), Sjödahl (n = 29), Stransky (n = 22), and TCGA (n = 404) data; exome profiles (n = 388) and FGFR3 and PPARG fusion data (n = 404) from TCGA data; CDKN2A and RB1 MLPA data from CIT (n = 86 and n = 85, respectively) and Stransky (n = 16 and n = 13, respectively) data; FGFR3 mutation data from MDA (n = 66), CIT (n = 87), Iyer (n = 39), Sjödahl (n = 28), and Stransky (n = 35); TP53 mutation data from MDA (n = 66), CIT (n = 87), Iyer (n = 39), Sjödahl (n = 28), and Stransky (n = 19); and RB1 mutation data from MDA (n = 66), CIT (n = 85), Iyer (n = 39), and Stransky (n = 13). Lower panels: associations between each consensus class, each type of gene alteration, and the combined alterations were evaluated by Fisher’s exact test. Consensus classes significantly enriched with alterations of these candidate genes are marked with a black asterisk.
Ba/Sq = basal/squamous; CIT = Cartes d’Identité des Tumeurs; CNA = copy number aberration; LumNS = luminal nonspecified; LumP = luminal papillary; LumU = luminal unstable; MDA = MD Anderson Cancer Center; MIBC = muscle-invasive bladder cancer; NE = neuroendocrine; TCGA = the Cancer Genome Atlas.
