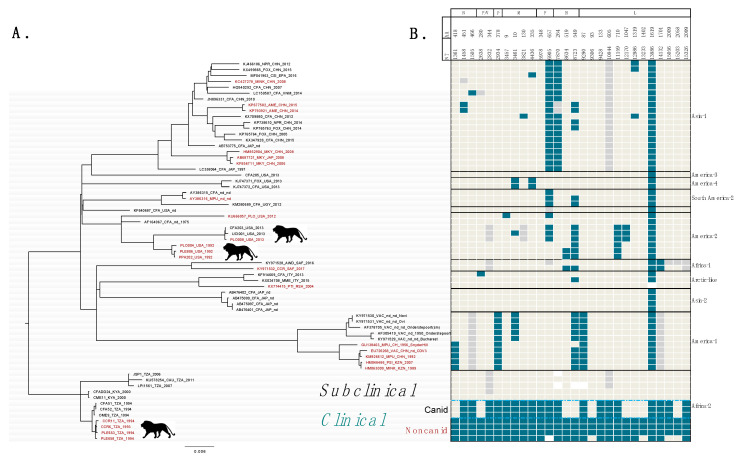
Figure 2.
Phylogenetic relationships among globally distributed canine distemper virus sequences and distribution of candidate markers of CDV pathogenicity in African lions. (A) Maximum likelihood (ML) phylogenetic tree of near-whole CDV genomes (15,584 bp) generated in this study and publicly available sequences collected globally. ML tree is rooted on the Africa-2 clade. Black circles at tree tips indicate sequences generated for this study. Font color of sequences, black, red, or grey indicates canid, non-canid, or vaccine origin, respectively. Clades implicated in distemper clinical signs in African lions indicated by lion icon. Bootstrap support of 500 replicates shown at nodes (B) SNPs at candidate loci are mapped to tips of the phylogeny representing globally distributed CDV strains. The genomic position of SNPs at candidate markers are shown in top row. Font color indicates whether overall selection at site is episodic diversifying (orange) or purifying (black). Teal blocks indicate identity with the African lion clinical strain circa Serengeti 1993–1994, taupe blocks = conserved allele, grey = alternative allele, white = no data.