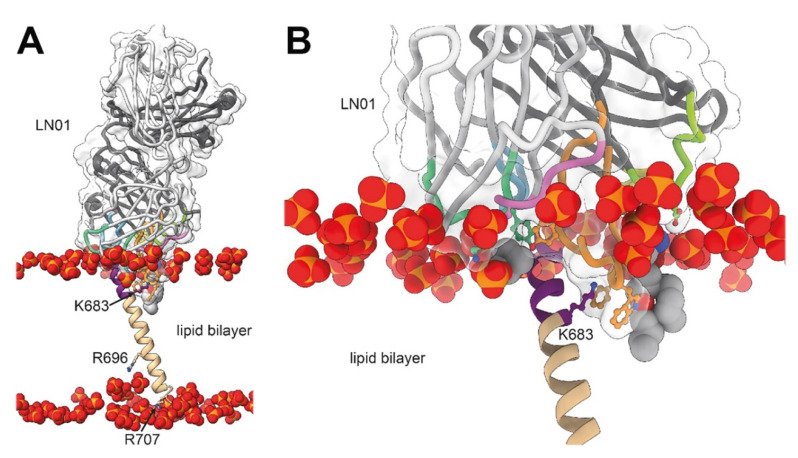
Figure 3.
The orientation of the MPER-TM epitope bound to LN01 in the lipid bilayer. (A,B) Representation of the LN01-MPER-TM complex placed into a bilayer with a lipid composition resembling the viral envelope obtained by molecular dynamics (MD) simulations. The LN01-MPER-TM complex and lipid analogues are represented as shown in Figure 1A and Figure 2A. Only the phosphate group of the lipids of the bilayer are displayed and represented as spheres. (A) The tilted orientation of the TM segment with respect to that of the bilayer allows residue K683 to interact with lipid head groups of one leaflet and R696 to contact head groups of the opposite leaflet; residues R707 and R709 terminating the TM helix interact as well with lipid head groups in the simulation. (B) Close up of the interaction between LN01, gp41 and the lipids. The gp41 residues located on the same side of the MPER-TM helix as K683 are more accessible to the interaction with the antibody. This orientation of the TM in the lipid bilayer could explain why only one side of the MPER-TM helical epitope (residues 673–686) is targeted by all known MPER bnAbs. The MD simulation supports the interaction of the Fab surface with the bilayer and underlines together with specific lipid-binding shown in later Figures, the recognition of a bipartite epitope composed of MPER and the membrane by MPER bnAbs.