Abstract
Emerging evidence indicates that microRNAs play a pivotal role in neural remodeling after spinal cord injury (SCI). This study aimed to investigate the mechanisms of miR-135a-5p in regulating the functional recovery of SCI by impacting its target genes and downstream signaling. The gene transfection assay and luciferase reporter assay confirmed the target relationship between miR-135a-5p and its target genes (specificity protein 1 [SP1] and Rho-associated kinase [ROCK]1/2). By establishing the H2O2-induced injury model, miR-135a-5p transfection was found to inhibit the apoptosis of PC12 cells by downregulating the SP1 gene, which subsequently induced downregulation of pro-apoptotic proteins (Bax, cleaved caspase-3) and upregulation of anti-apoptotic protein Bcl-2. By measuring the neurite lengths of PC12 cells, miR-135a-5p transfection was found to promote axon outgrowth by downregulating the ROCK1/2 gene, which subsequently caused upregulation of phosphate protein kinase B (AKT) and phosphate glycogen synthase kinase 3β (GSK3β). Use of the rat SCI models showed that miR-135a-5p could increase the Basso, Beattie, and Bresnahan (BBB) scores, indicating neurological function recovery. In conclusion, the miR-135a-5p-SP1-Bax/Bcl-2/caspase-3 and miR-135a-5p-ROCK-AKT/GSK3β axes are involved in functional recovery of SCI by regulating neural apoptosis and axon regeneration, respectively, and thus can be promising effective therapeutic strategies in SCI.
Keywords: spinal cord injury, PC12 cell, microRNA-135a-5p, SP1, ROCK
Graphical Abstract
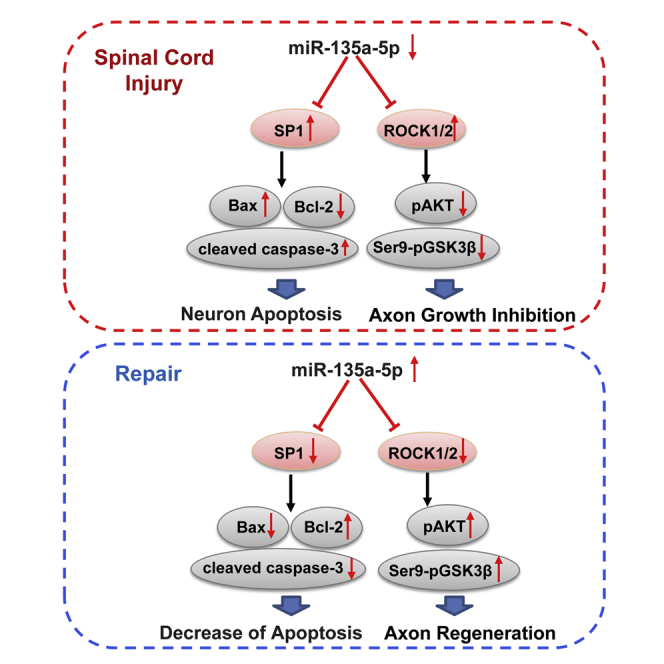
Wang et al. demonstrate that upregulating miR-135a-5p can promote axon outgrowth by modulating the ROCK-AKT/GSK3β pathway and suppress neural apoptosis by inhibiting the SP1-Bcl-2/Bax/caspase-3 pathway. These findings provide two novel signaling axes (miR-135a-5p-ROCK-AKT/GSK3β and miR-135a-5p-SP1-Bcl-2/Bax/caspase-3) that may be potential therapeutic targets in promoting functional recovery following spinal cord injury.
Introduction
Spinal cord injury (SCI) is a severe central nervous system disease that can cause permanent disabilities. However, there is currently no effective therapy used in the clinic.1 It is generally recognized that SCI-induced neurological deficits are not only a consequence of the initial damage to the parenchyma, but also the secondary events that occur after the primary injury, which increase the complexity and refractory nature of the disease.2 Oxidative stress, caused by reactive oxygen species (ROS), following SCI plays a crucial role in cell death and lesion expansion and can lead to the apoptosis of neurons.3, 4, 5 Alternatively, axon regrowth can be inhibited by neuronal growth inhibitors in the local environment after SCI.6 Therefore, investigating the genetic and epigenetic mechanisms of neuronal protection and axon regrowth has been the focus of studies on SCI repair.7,8 MicroRNAs (miRNAs) can bind to the 3′ untranslated region (UTR) of target messenger RNAs and, thus, negatively regulate gene expression at the mRNA and protein levels.9 Increasing numbers of studies have shown the regulation of miRNAs in promoting axon outgrowth and inhibiting oxidative stress-induced neuronal apoptosis.10, 11, 12
miR-135a-5p has been verified to be downregulated in injured spinal cord tissue by analyzing the Gene Expression Omnibus (GEO): GSE19890 dataset. It was found to be a stimulator of axon regrowth and also play a neuroprotective role by inhibiting cellular apoptosis.13,14 However, there is still no report investigating the mechanisms of miR-135a-5p in regulating the neuronal functional recovery of SCI, especially in terms of its target genes and the signaling pathway. Based on the predicted results of the TargetScan searching tool,15 635 genes were found to be the targets of miR-135a-5p. Among these 635 genes, specificity protein 1 (SP1) and Rho-associated kinase (ROCK) seem to be important since they were implicated in regulating neural apoptosis and axon regeneration.16, 17, 18, 19 It is also known that the target genes obtained from TargetScan should be determined based on either experimental verification or subsequent computational prediction, namely requiring confirmation by performing the luciferase reporter assay in the present study. The results of this assay identified the binding sequence of the target sites between miR-135a-5p and these two genes and, thus, validated their target relationships. Based on the results obtained from the TargetScan searching tool and our present study, SP1 and ROCK1/2 were thought to have great significance in SCI repair and hypothesized to be involved in miR-135a-5p-associated axes in mediating neuronal apoptosis and axon regeneration.
SP1 was found to be consistently overexpressed in motor neurons during the injury response following SCI.20 The overexpression of SP1 induced by neuronal oxidative stress was shown to induce cell apoptosis by binding several apoptosis-related genes (Bax, Bcl-2, and caspase-3) and activating the apoptosis pathway consisting of the Bax/Bcl-2/caspase-3 axis.16,21,22 The downregulation of Bcl-2, as well as the upregulation of Bax and caspase-3, have been detected in injured spinal cord tissue.23,24 Based on these facts, SP1 and its downstream Bax/Bcl-2/caspase-3 signaling axis were chosen for further investigation regarding the mechanism of miR-135a-5p in regulating neuronal apoptosis. To date, there have been no reports clarifying the relationship between miR-135a-5p and Sp1 in neuronal protection following SCI.
Rho-associated kinase (ROCK) has been confirmed as a direct target gene of miR-135a-5p in multiple diseases.25,26 Since ROCK can act as an axon growth inhibitor, the inhibition of ROCK has been shown to be a potential therapeutic approach to enhance axon outgrowth.27 Moreover, the phosphate protein kinase B (AKT)/glycogen synthase kinase 3β (GSK3β) pathway was shown to be a potential downstream signaling pathway of ROCK, since ROCK activity activates phosphatase and tensin homolog (PTEN) to downregulate Akt activity and upregulate GSK3β activity in sequence.28 The activation of AKT and the GSK3β pathway has been shown to regulate the neuron growth by promoting the establishment, elongation, and development of axons.29,30 Based on these facts, ROCK and its downstream AKT/GSK3β pathways were selected for further investigation regarding the mechanisms of miR-135a-5p in regulating axon regeneration.
This study aimed to investigate the mechanisms of miR-135a-5p in regulating neuronal apoptosis and axon regeneration following SCI. The target relationship between miR-135a-5p and genes (SP1 and ROCK) was found to be involved in neuronal apoptosis and axon outgrowth during the neural recovery process following SCI. The miR-135a-5p/Sp1 and miR-135a-5p/ROCK axes might be promising therapeutic targets to promote the functional recovery of damaged spinal cords.
Results
Identification of Differentially Expressed miRNAs Based on a Selected Public Dataset
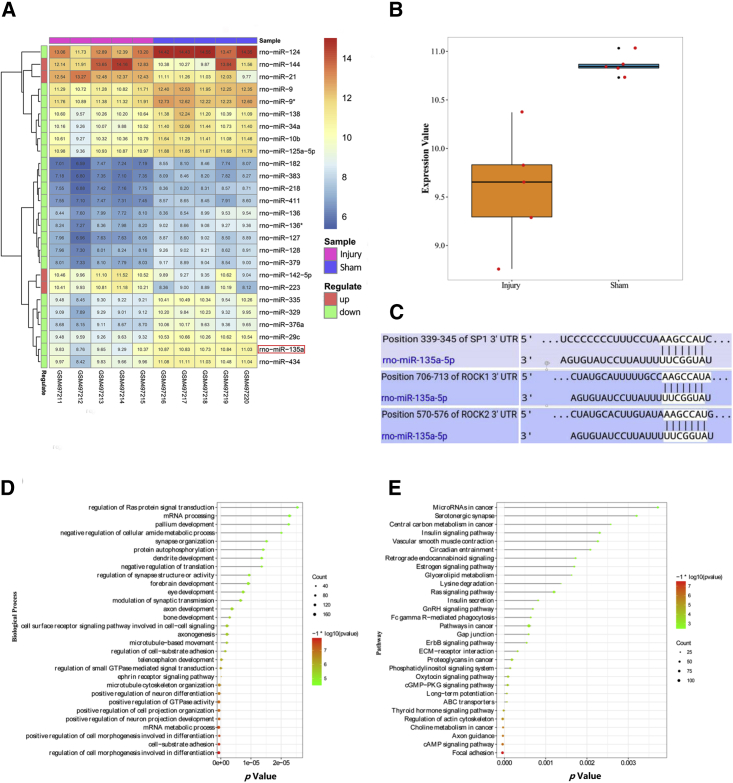
As shown in Figure 1A, 26 differentially expressed (DE) miRNAs were identified by differential expression analysis. This included 4 upregulated DE miRNAs (miR-223, miR-144, miR-21, and miR-142-5p) and 22 downregulated DE miRNAs (e.g., miR-434, miR-135a, miR-29c, and miR-376a).
Figure 1.
miR-135a-5p Expression Levels Were Decreased after SCI, and miR-135a-5p May Target SP1 and ROCK1/2 and Modulate Neurological Recovery
(A) Heatmap of the differentially expressed miRNAs. (B) rno-miR-135a was significantly downregulated after injury. (C) TargetScan prediction of target sites between miR-135a-5p and SP1 and ROCK1/2. (D and E) Enriched biological processes (D) and signaling pathways (E) of the target genes of miR-135a-5p, rno-miR-135a, and miR-135a-5p.
Functional Similarity between Literature-Mined miRNAs and DE miRNAs with a High Correlation
By mining the scientific literature,31, 32, 33, 34 32 miRNAs were identified to be closely related to neurological recovery. After Pearson’s correlation coefficient (PCC) analysis, 10 miRNAs were found to be highly associated with neurological recovery. Table 1 shows the functional similarity between the 32 miRNAs identified by literature mining and the 10 miRNAs identified by PCC analysis. miR-135a-5p and miR-124-3p were highly similar, and their expression levels were correlated. miR-135a-5p had a significant advantage in terms of all parameters, including the Jaccard index pathway (JI path), Jaccard index biological processes (JI bps), Jaccard index gene targets (JI targets), and the correlation coefficient (CC). miR-124-3p is an experimentally validated miRNA, with a role in promoting neurological recovery.35, 36, 37 The target genes of miR-135a-5p were closely associated with the signaling pathways of neurological recovery. All of these analyses showed that miR-135a-5p was significantly downregulated after injury (Figure 1B). These findings showed that miR-135a-5p dysregulation might be involved in SCI by regulating neurological function recovery.
Table 1.
The Functional Similarity Relationship between the 32 miRNAs Obtained by Literature Mining and 10 miRNAs Obtained by the PCC Analysis
| Known miRNA | DE miRNA | JI Path | JI bps | JI Targets | CC |
|---|---|---|---|---|---|
| rno-miR-124-3p | rno-miR-135a-5p | 0.3372 | 0.1175 | 0.2431 | 0.9351 |
| rno-miR-124-3p | rno-miR-379-5p | 0.3218 | 0.0843 | 0.2126 | 0.95673 |
| rno-miR-26a-5p | rno-miR-127-3p | 0.2812 | 0.1458 | 0.26 | 0.96613 |
| rno-miR-124-3p | rno-miR-411-5p | 0.2609 | 0.1106 | 0.4526 | 0.97167 |
| rno-miR-124-3p | rno-miR-411-5p | 0.2239 | 0.0391 | 0.1531 | 0.96323 |
| rno-miR-541-5p | rno-miR-383-5p | 0.2222 | 0.0942 | 0.2582 | 0.96027 |
| rno-miR-124-3p | rno-miR-10b-5p | 0.1935 | 0.127 | 0.1954 | 0.97697 |
| rno-miR-182 | rno-miR-128-3p | 0.1772 | 0.3117 | 0.3238 | 0.97397 |
| rno-miR-34a-5p | rno-miR-434-3p | 0.1429 | 0.1658 | 0.2019 | 0.95197 |
| rno-miR-34a-5p | rno-miR-329-3p | 0.1429 | 0.1349 | 0.3789 | 0.97873 |
| rno-miR-124-3p | rno-miR-434-3p | 0.1343 | 0.0843 | 0.2423 | 0.954 |
| rno-miR-124-3p | rno-miR-383-5p | 0.1186 | 0.1675 | 0.3438 | 0.9617 |
| rno-miR-182 | rno-miR-125a-5p | 0.1053 | 0.1591 | 0.243 | 0.97253 |
| rno-miR-34a-5p | rno-miR-329-3p | 0.0909 | 0.0411 | 0.1463 | 0.95967 |
| rno-miR-124-3p | rno-miR-379-5p | 0.0702 | 0.2059 | 0.2541 | 0.9807 |
| rno-miR-182 | rno-miR-379-5p | 0.0588 | 0.0946 | 0.2274 | 0.95337 |
JI, Jaccard index, which means the similarity between the two miRNAs; Path, pathway; bps, biological processes; CC, correlation coefficient.
Biological Processes and Signaling Pathways Enriched for miR-135a-5p
The enriched biological processes and signaling pathways of the target genes of miR-135a-5p are shown in Figures 1D and 1E, respectively. As shown in Figure 1D, the target genes of miR-135a-5p were involved in several biological processes related to neural regeneration, such as axon development, microtubule-based movement, microtubule cytoskeleton organization, positive regulation of neuron differentiation, and positive regulation of neuron projection development. As shown in Figure 1E, the target genes of miR-135a-5p were involved in several signaling pathways related to neural regeneration, such as insulin signaling, estrogen signaling, Ras signaling, ErbB signaling, actin cytoskeleton regulation, axon guidance, and focal adhesion. Based on these findings, we speculated that miR-135a-5p played a critical role in functional recovery after SCI.
Relationship between miR-135a-5p and SP1 and ROCK1/2
As shown in Figure 1C, miR-135a-5p targeted position 339–345 of the 3′ UTR of SP1, position 706–713 of the 3′ UTR of ROCK1, and position 570–576 of the 3′ UTR of ROCK2. Based on the fact that miRNAs negatively regulate the expression of their target genes, the downregulation of miR-135a-5p was predicted to induce the upregulation of SP1 and ROCK1/2 genes following SCI,38,39 thus validating the negative relationship between the expression of miR-135a-5p and SP1 and ROCK1/2.
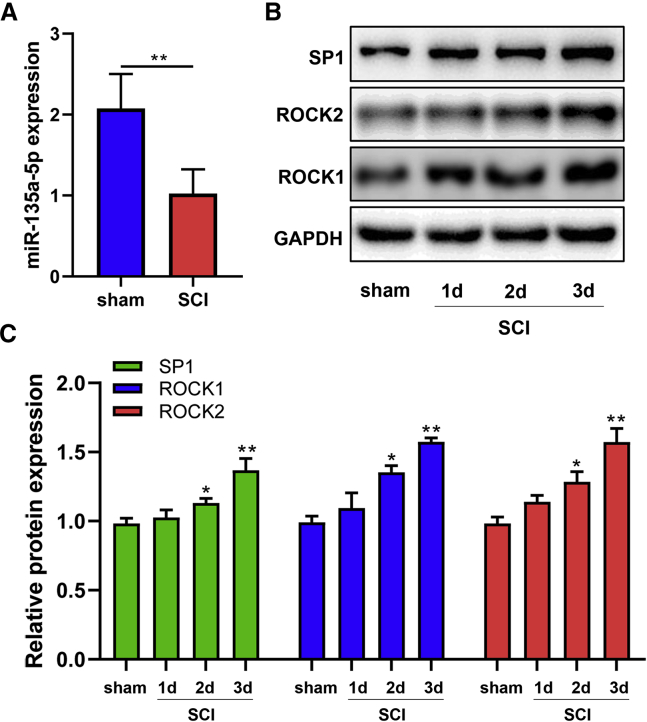
Expression of miR-135a-5p, SP1, and ROCK after SCI
Rat SCI models were established to verify the aforementioned bioinformatics results. The results demonstrated that the miR-135a-5p expression level was significantly decreased at 3 days after SCI when compared with those in the sham group (Figure 2A). Moreover, the expression levels of SP1, ROCK1, and ROCK2 proteins were significantly increased compared with their levels in the sham group, 3 days after SCI (Figures 2B and 2C).
Figure 2.
The Expression Levels of miR-135a-5p Were Reduced, while SP1 and ROCK1/2 Were Overexpressed in the Damaged Spinal Cord Tissues of Rats
Spinal cord samples were collected and analyzed. (A) miR-135a-5p was significantly decreased at 3 days post-SCI (n = 3). (B) Protein levels in spinal cords. (C) The expression levels of SP1, ROCK1, and ROCK2 proteins on 2 and 3 days post-SCI were significantly increased compared with that examined in the sham group (n = 3). Statistical data were measurement data, described as mean ± standard deviation. ∗p < 0.05, ∗∗p < 0.01, when compared with the sham group.
H2O2 Induced PC12 Cell Injury
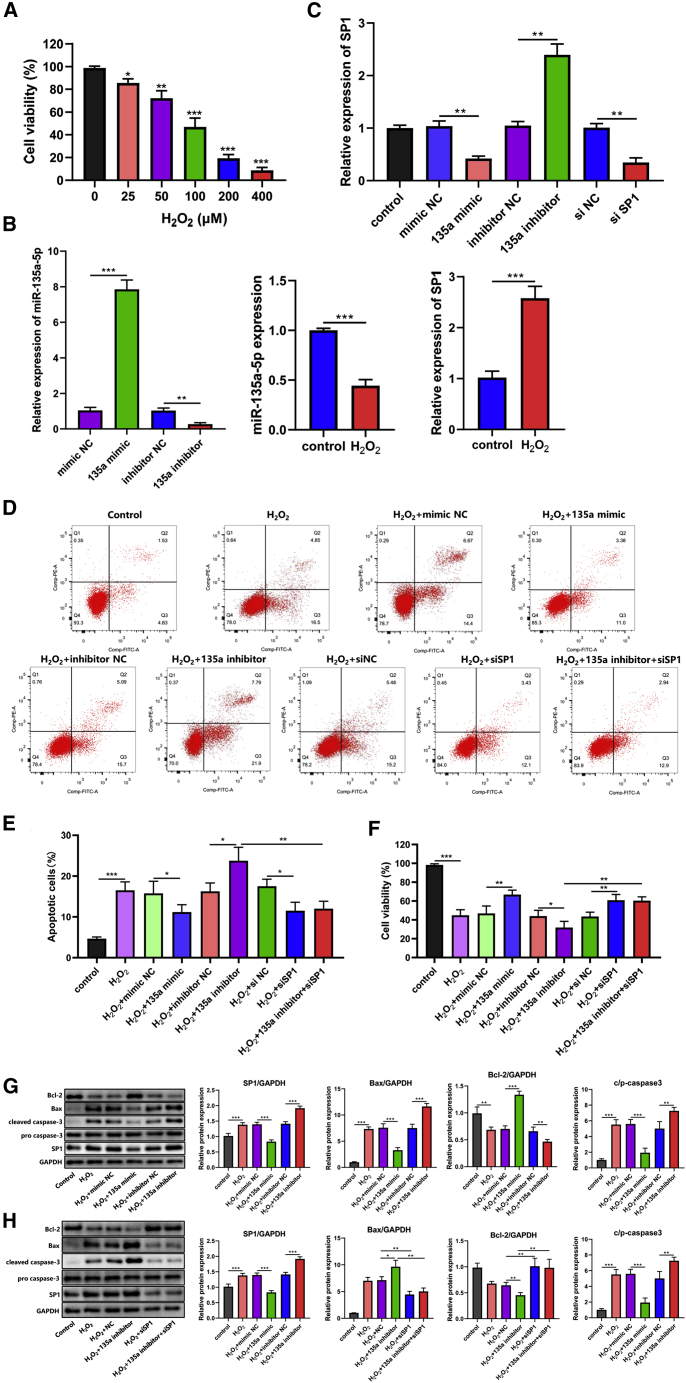
Different dosages of H2O2 (0-400 μM) were used to stimulate PC12 cells, and cell viability was subsequently determined using the Cell Counting Kit-8 (CCK-8) assay. PC12 cell viability was clearly reduced by H2O2 at doses of 25, 50, 100, 200, and 400 μM. Based on the fact that 100 μM H2O2 suppressed cell viability by 50% (Figure 3A), this dosage was used in subsequent experiments to stimulate PC12 cells. Apoptosis assays showed that the percentage of apoptotic cells was markedly higher in the H2O2-treated group than in the control group (Figures 3D and 3E). Similarly, the western blotting analysis showed that H2O2 downregulated Bcl-2 (anti-apoptotic protein) levels and upregulated Bax and cleaved caspase-3 (pro-apoptotic factors) levels, resulting in increased apoptosis (Figures 3G and 2H). These results suggested that H2O2 promoted the apoptosis of PC12 cells.
Figure 3.
miR-135a-5p Protects against H2O2-Induced PC12 cell Damage
(A) PC12 cells were treated with various H2O2 dosages (0–400 μM), and CCK-8 experiments were performed to assess cell viability. The dosage of 100 μM H2O2 was selected and used to treat PC12 cells (n = 6). (B) Expression levels of miR-135a-5p or SP1 in PC12 cells after miR-135a-5p transfection and H2O2 stimulation (n = 3). (C) RNA levels of SP1 in cells transfected with 135a mimic, 135a inhibitor, siSP1, or their NCs (n = 3). (D) Apoptotic cell rate (n = 6). (E) The percentage of apoptotic cells significantly increased in H2O2 group; The apoptotic rate decreased in cells transfected with miR-135a-5p mimics, compared to the NC group; the apoptotic rate was significantly increased in the miR-135a-5p inhibitor group compared with the NC group; The siSP1 could rescue the apoptosis induced by miR-135a-5p inhibitor. (F) Cell viability (n = 6). (G) The levels of pro-apoptotic proteins (cleaved caspase-3 and Bax) were downregulated, while significant increases in the levels of anti-apoptotic proteins (Bcl-2) were observed in miR-135a-5p mimic group or an siSP1 group; whereas Bax and cleaved caspase-3 were upregulated, Bcl-2 was downregulated in the miR-135a-5p inhibitor group. (n = 3). (H) The downregulation of SP1 rescued the apoptosis rate and apoptosis-related protein levels that were decreased by the miR-135a-5p inhibitor. (n = 3). Statistical data were measurement data, described as mean ± standard deviation; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NC, negative control; 135a, miR-135a-5p; siSP1, siRNA targeting SP1.
Overexpression of miR-135a-5p Improved PC12 Cell Viability and Reduced Apoptosis
The expression levels of miR-135a-5p or SP1 in PC12 cells were determined by using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis after miR-135a-5p transfection and H2O2 stimulation. The results showed that PC12 cells were stably transfected with a miR-135a-5p mimic, a miR-135a-5p inhibitor, or their respective negative control (NC) RNAs (Figure 3B). miR-135a-5p expression levels were significantly decreased, while SP1 expression levels were significantly increased when compared with those in the control group at 24 h after H2O2 stimulation (Figure 3B). The effects of miR-135a-5p expression on H2O2-diminished cell viability were detected in PC12 cells. The viability of PC12 cells was found to be significantly decreased after H2O2 exposure. The viability of cells transfected with miR-135a-5p mimics was much higher than the viability of cells in the miR-135a-5p NC group, while cell viability was lower in the miR-135a-5p inhibitor group than in the miR-135a-5p NC group. Furthermore, the downregulation of SP1 fully rescued cell viability that was decreased by the miR-135a-5p inhibitor (Figure 3F).
The impact of miR-135a-5p on H2O2-induced apoptosis was then evaluated by assessing apoptosis severity and measuring changes in the expression of apoptosis-associated proteins following transfection. The percentage of apoptotic cells significantly increased following H2O2 exposure. The apoptotic rate decreased in cells transfected with miR-135a-5p mimics, compared to cells in the NC group that were treated with H2O2 (Figures 3D and 3E). In contrast, the apoptotic rate was significantly increased in the miR-135a-5p inhibitor group compared with the inhibitor NC group (Figures 3D and 3E). Furthermore, the western blotting analysis showed that, in the miR-135a-5p group, the levels of Bax and cleaved caspase-3 were lower, while the level of Bcl-2 was higher compared with the mimic NC group (Figure 3G). As expected, Bax and cleaved caspase-3 were upregulated, whereas Bcl-2 was downregulated in the miR-135a-5p inhibitor group when compared with the NC group (Figure 3G). These data suggested that miR-135a-5p overexpression protects PC12 cells against H2O2-induced apoptosis.
miR-135a-5p Protected PC12 Cells from Oxidative Damage by Inhibiting SP1 Expression
PC12 cells stably transfected with a miR-135a-5p mimic, a miR-135a-5p inhibitor, or their respective NC RNAs were treated with H2O2, after which SP1 expression and cellular injury were evaluated. The results showed that there was a significant downregulation of SP1 levels in the H2O2 + mimic cells when compared with the H2O2 + mimic NC cells, while there was a significant upregulation of SP1 levels in the H2O2 + inhibitor cells compared with the H2O2 + inhibitor NC cells. Furthermore, small interfering RNA (siRNA) transfection successfully downregulated SP1 (Figure 3C).
However, H2O2-induced PC12 cell apoptosis was reversed by transfection with miR-135a-5p mimics or an SP1-specific siRNA (Figure 3H). When compared with the normal group, the levels of pro-apoptotic proteins (cleaved caspase-3 and Bax) were downregulated, while significant increases in the levels of anti-apoptotic proteins (Bcl-2) were observed (Figures 3G and 3H). Meanwhile, lower expression levels of anti-apoptotic proteins and higher expression levels of pro-apoptotic proteins were found in the H2O2 + miR-135a-5p inhibitor group compared with the H2O2 + inhibitor NC group (Figure 3G). Furthermore, the downregulation of SP1 fully rescued the apoptosis rate and apoptosis-related protein levels that were decreased by the miR-135a-5p inhibitor (Figure 3H).
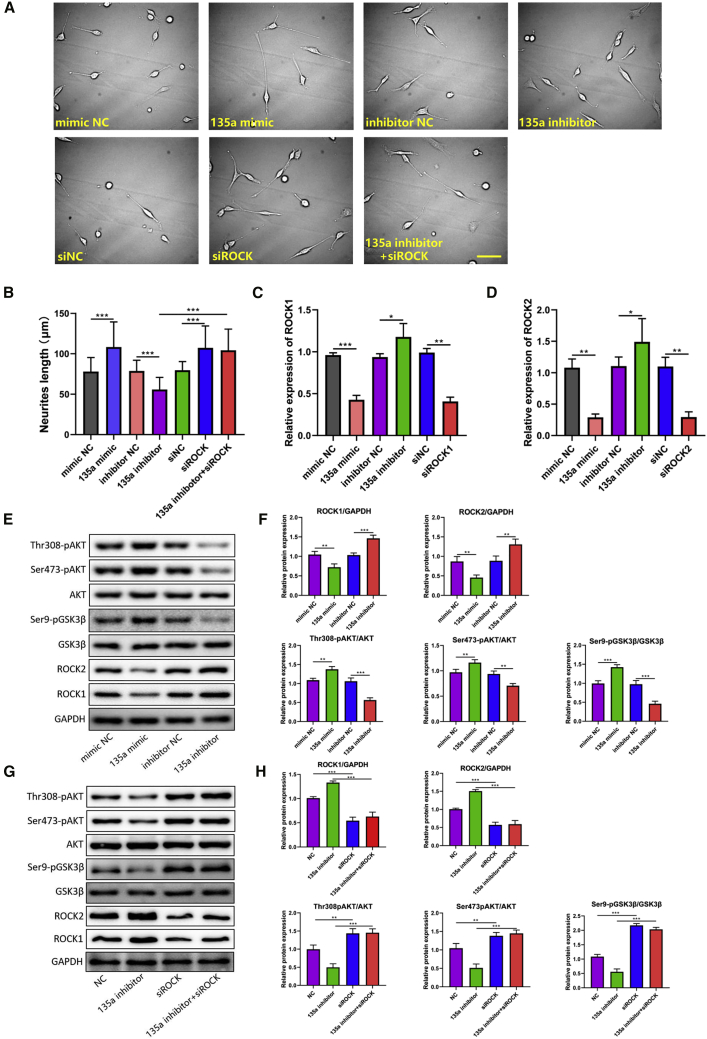
miR-135a-5p Promoted Neurite Outgrowth in PC12 Cells by Inhibiting ROCK Expression and Regulating the AKT/GSK3β Signaling Pathway
In order to investigate whether miR-135a-5p can regulate neurite outgrowth, PC12 cells were transfected with miR-135a-5p mimic, miR-135a-5p inhibitor, or their respective NC RNAs. The final results showed that axons in PC12 cells transfected with the miR-135a-5p mimic had a significant increase in neurite extension compared to those transfected with the NC mimic (Figures 4A and 4B). Furthermore, whether the knockdown of miR-135a-5p could inhibit the neurite extension was examined in the present study. PC12 cells transfected with the miR-135a-5p inhibitor showed a significant decrease in neurite length (Figures 4A and 4B).
Figure 4.
Overexpression of miR-135a-5p Promotes Neurite Outgrowth in PC12 Cells
(A) Neurite length in PC12 cells transfected with 135a mimic, 135a inhibitor, siROCK, or their NC (scale bar, 100 μm; n = 100). (B) The PC12 cells transfected with the miR-135a-5p mimic group had a significant increase in neurite extension compared to those transfected with the NC group; Cells transfected with the miR-135a-5p inhibitor showed a significant decrease in neurite length (n = 100). (C and D) RNA levels of ROCK1 (C) and ROCK2 (D) after siRNA transfection in cells (n = 3). (E) Protein levels of ROCK1/2, Ser473-pAKT/AKT, Thr308-pAKT/AKT, and Ser9-pGSK3β/GSK3β (n = 3). (F) The expression levels of ROCK1 and ROCK2 were significantly decreased in miR-135a-5p mimic group, whereas the miR-135a-5p inhibitor group increased ROCK1 and ROCK2 levels; The levels of pAKT (Ser473 and Thr308) and pGSK3β (Ser9) were significantly higher in miR-135a-5p mimic group; Moreover, the miR-135a-5p inhibitor group reduced the levels of pAKT (Ser473 and Thr308) and pGSK3β (Ser9) (n = 3). (G) Protein levels of ROCK1/2, Ser473-pAKT/AKT, Thr308-pAKT/AKT, and Ser9-pGSK3β/GSK3β (n = 3). (H) the reduced levels of pAKT (Ser473 and Thr308) and pGSK3β (Ser9) induced by the miR-135a-5p inhibitor were rescued by the downregulation of ROCK (n = 3). Statistical data were measurement data, described as mean ± standard deviation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NC, negative control; 135a, miR-135a-5p; siROCK, siRNA targeting ROCK1 and siRNA targeting ROCK2.
qRT-PCR analysis demonstrated that the mRNA levels of ROCK1 and ROCK2 were affected by the modulation of miR-135a-5p and by transfection with ROCK1- and ROCK2-specific siRNAs (Figures 4C and 4D). In addition, western blotting analysis indicated that the expression levels of ROCK1 and ROCK2 were significantly decreased in PC12 cells transfected with the miR-135a-5p mimic, whereas inhibitors of miR-135a-5p increased ROCK1 and ROCK2 levels (Figures 4E and 4F). Afterward, the underlying mechanism of miR-135a-5p-ROCK in regulating axon outgrowth was explored by investigating its potential downstream AKT/GSK3β pathway. The experimental results showed that the phosphorylation levels of AKT (Ser473 and Thr308) and GSK3β (Ser9) were significantly higher in miR-135a-5p-overexpressing cells than that in cells expressing the NC mimic. Moreover, cells treated with miR-135a-5p inhibitor showed reduced phosphorylation levels of members of this pathway compared to cells in the inhibitor NC group (Figures 4E and 4F). These results validated that miR-135a-5p dysregulation impacted AKT/GSK3β signaling.
The Regulatory Effect of miR-135a-5p on Axon Outgrowth and the AKT/GSK3β Signaling Pathway Was Mediated by ROCK
To determine whether miR-135a-5p inhibition impaired axon regeneration by regulating ROCK1 and ROCK2 levels, rescue experiments were performed in order to examine whether the downregulation of ROCK1 and ROCK2 was able to reverse the inhibitory effect of the miR-135a-5p inhibitor on axon outgrowth. The downregulation of ROCK1 and ROCK2 was found to fully rescue the axon regrowth that was blocked by the miR-135a-5p inhibitor (Figures 4A and 4B). Similarly, the reduced levels of phosphorylated AKT (Ser473 and Thr308) and GSK3β (Ser9) induced by the miR-135a-5p inhibitor were reversed by the downregulation of ROCK1 and ROCK2 (Figures 4G and 4H). These results showed that dysregulation of the miR-135a-5p-regulated AKT/GSK3β pathway was mediated by ROCK1/2.
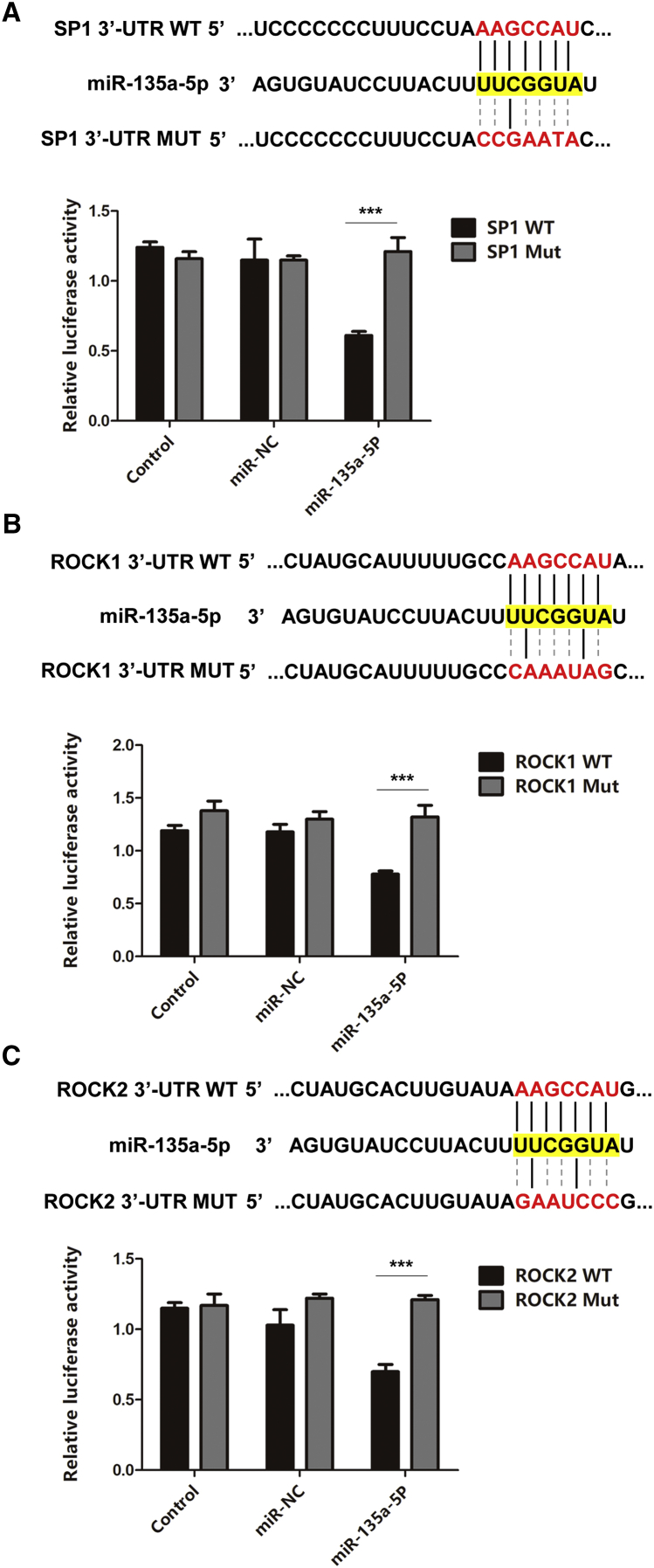
miR-135a-5p Directly Targeted SP1, ROCK1, and ROCK2
The luciferase reporter assay was performed to confirm the binding relationship between miR-135a-5p and three potential target genes (SP1, ROCK1, and ROCK2). As expected, miR-135a-5p mimics markedly inhibited the luciferase activity of wild-type (WT) SP1, ROCK1, and ROCK2 reporters in HEK293T cells. However, luciferase activity of mutant (MUT) SP1, ROCK1, and ROCK2 reporters was not significantly changed following miR-135a-5p mimic or NC mimic transfection (Figures 5A–5C). These findings indicated that miR-135a-5p directly targeted SP1, ROCK1, and ROCK2.
Figure 5.
Identification of SP1, ROCK1, and ROCK2 as Target Genes of miR-135a-5p
Potential target sites of miR-135a-5p on the mRNA 3′ UTRs and several point mutations were generated at the binding site. A luciferase reporter assay identified the predicted binding sequence required for miR-135a-5p-mediated inhibition. (A) SP1 luciferase assay (n = 3). (B) ROCK1 luciferase assay (n = 3). (C) ROCK2 luciferase assay (n = 3). Statistical data were measurement data, described as mean ± standard deviation. ∗∗∗p < 0.001. NC, negative control.
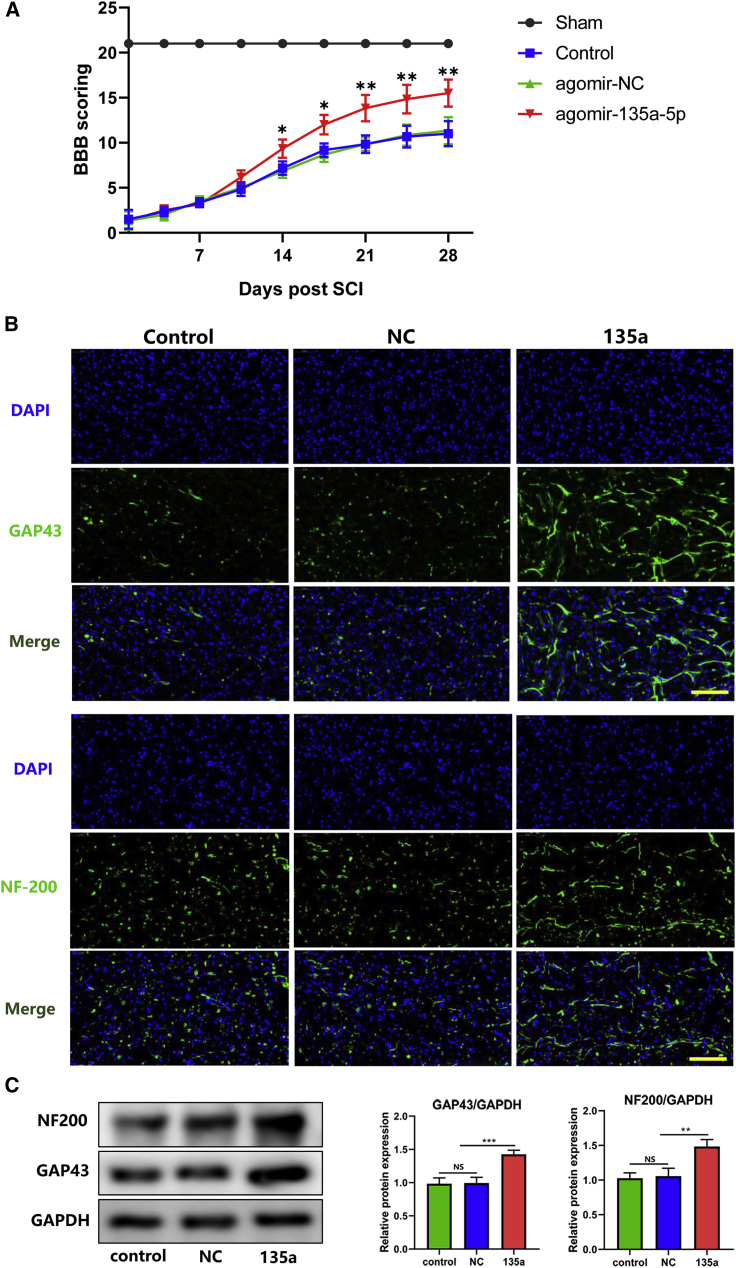
miR-135a-5p Promoted Functional Recovery and Neurite Outgrowth after SCI
Functional recovery after SCI was further analyzed, and the data demonstrated that miR-135a-5p significantly increased Basso, Beattie, and Bresnahan (BBB) scores after SCI, compared with those of the control and NC groups, and that the BBB scores further increased 4 weeks after treatment (Figure 6A).
Figure 6.
Functional Recovery of Damaged Spinal Cords after Treatment of agomir-135a-5p
(A) Motor recovery of the hind limb was monitored after SCI using Basso, Beattie, and Bresnahan scoring (n = 6). (B) GAP43 and NF200 expression levels were measured by immunofluorescence staining (scale bars, 100 μm; n = 3). (C) Protein levels of GAP43 and NF200 (n = 3). Statistical data were measurement data, described as mean ± standard deviation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant (p > 0.05); NC, negative control; 135a, miR-135a-5p.
Immunofluorescence analysis showed that miR-135a-5p promoted the expression of growth-associated protein 43 (GAP43) and neurofilament 200 (NF-200) at day 7 after intrathecal injection, while the control group and the agomir-NC group, with no miR-135a-5p treatment, showed lower levels of expression of these two markers, indicating impaired axon regeneration (Figure 6B). In addition, GAP43 and NF-200 protein levels were measured using western blotting. According to these data, the expression levels of GAP43 and NF-200 increased in the group treated with miR-135a-5p (Figure 6C).
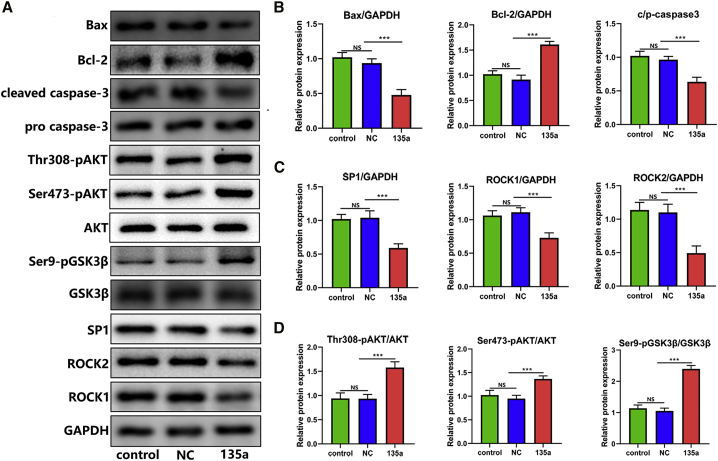
miR-135a-5p Inhibited Apoptosis after SCI
Apoptosis levels were evaluated in injured spinal cords. Bcl-2, Bax, and caspase-3 levels were measured. The expression levels of Bcl-2 were found to be markedly increased, and the levels of Bax and caspase-3 were reduced in the agomir-miR-135a-5p treatment group (Figures 7A and 7B). This suggested that high levels of miR-135a-5p mitigated secondary injury and, thus, downregulated pro-apoptotic protein levels in the damaged spinal cord. In summary, miR-135a-5p mimic treatment suppressed apoptosis in SCI.
Figure 7.
Protein Levels in Injured Spinal Cords after Treatment with miR-135a-5p
(A) Western blotting analysis was performed to indicate the expression levels of proteins in injured spinal cords. (B) miR-135a-5p increased the levels of Bcl-2 and decreased the levels of Bax, cleaved caspase-3. (C) miR-135a-5p decreased the levels of SP1, ROCK1, and ROCK2, and (D) promoted the phosphorylation of Ser473-AKT, Thr308-AKT, and Ser9-GSK3β in injured spinal cords. Statistical data were measurement data, described as mean ± standard deviation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. NS, not significant (p > 0.05); NC, negative control; 135a, miR-135a-5p.
miR-135a-5p Decreased the Levels of SP1 and ROCK, while It Increased the Levels of Phosphorylated AKT and Its Targets in the Injured Spinal Cord
Western blotting analysis indicated that the expression levels of SP1, ROCK1, and ROCK2 were significantly decreased in the agomir-miR-135a-5p group (Figures 7A and 7C). The western blotting analysis was also performed to assess whether the increase in miR-135a-5p level was upstream of the activated AKT pathway after SCI. These results demonstrated that miR-135a-5p treatment was highly associated with increased levels of phosphorylated AKT and the AKT substrate, GSK3β. Total AKT and GSK3β levels were unaltered after miR-135a-5p treatment. Notably, miR-135a-5p significantly upregulated the levels of activated (pSer473, pThr308) AKT and increased the phosphorylation and AKT-dependent inactivation of GSK3β (pSer9, Figures 7A and 7D). There were no significant differences between the control and agomir-NC groups.
Discussion
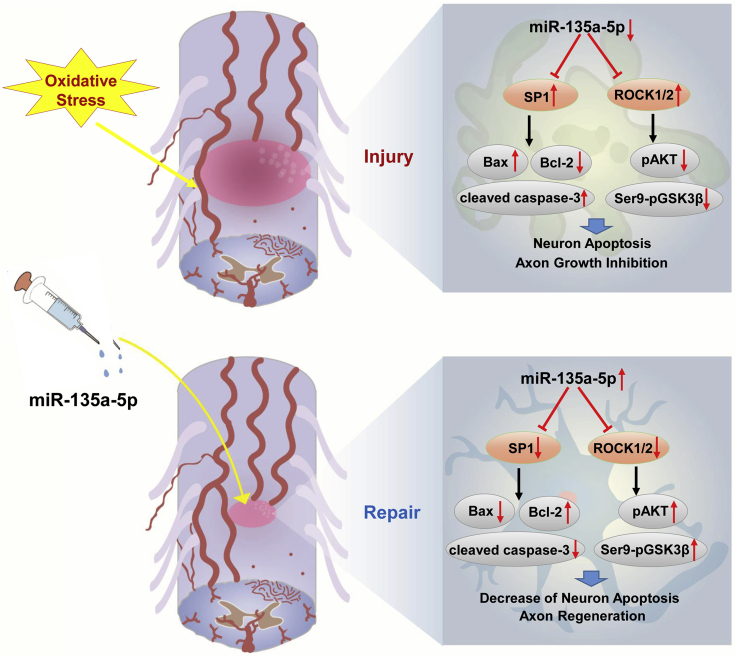
Using both in vitro and in vivo experiments, the present study implicated the miR-135a-5p-SP1-Bax/Bcl-2/caspase-3 axis in inhibiting neuronal apoptosis and the involvement of the miR-135a-5p-ROCK-AKT/GSK3β signaling pathway in promoting axon regeneration during the process of functional recovery following SCI. The mechanisms by which these two signaling axes are involved in regulating functional recovery following SCI may provide potential therapeutic targets for clinical applications (Figure 8).
Figure 8.
Proposed Mechanism of the Role of miR-135a-5p in SCI
The expression of miR-135a-5p can reduce apoptosis and increase axon outgrowth by inhibiting SP1 and ROCK, thus promoting the functional recovery of damaged spinal cords.
The miR-135a-5p-SP1 axis was found to be involved in the functional recovery of damaged spinal cord tissues by regulating apoptosis. The results of CCK-8 and flow cytometry assays (Figures 3D–3F) showed that the overexpression of miR-135a-5p inhibited H2O2-induced cell death and vice versa. In contrast to our findings, a study using the H2O2-induced injury model in rat cardiomyoblast cells showed that the overexpression of miR-135a-5p enhanced H2O2-induced apoptosis.40 These contradictory results may be due to different expression patterns and roles of miR-135a-5p in different tissues and cell types. Moreover, the present study verified the relationship between miR-135a-5p and SP1 since miR-135a-5p was found to significantly inhibit the expression of SP1 (Figures 3G and 5A). The present study is the first to identify the regulatory mechanism of miR-135a-5p and SP1 during the pathophysiology of SCI. Subsequently, the present study found that miR-135a-5p inhibitor-induced apoptosis was associated with the upregulation of SP1, indicating that SP1 upregulation may promote neuronal apoptosis. Previous studies have shown similar results, suggesting the role of SP1 as an apoptosis-related transcription factor.16,41, 42, 43, 44
The role of the miR-135a-5p-SP1 axis in inhibiting apoptosis was verified by the finding that Bcl-2 was upregulated, and Bax and cleaved caspase-3 were downregulated.21 Bcl-2 family genes, consisting of pro-apoptotic members (Bax) and anti-apoptotic members (Bcl-2), are known to be downstream targets of the transcription factor SP1.45 The proper regulation of Bax transcription was discovered to be regulated by the binding sites of SP1 and E-box.46 As an initiator of the apoptotic death pathway, Bax can induce apoptotic cell death by regulating cell-cycle progression through the G1/S checkpoint.47 The overexpression of Bax detected in the injured spinal neurons could lead to neuronal apoptosis following SCI,48,49 which is also in agreement with our findings showing the upregulation of Bax after inhibiting miR-135a-5p (Figure 3G). Bcl-2 family members have been shown to play a pivotal role in regulating cell death by acting as critical decision points in the apoptosis pathway.50 A previous study showed that the overexpression of Bcl-2 suppressed apoptosis following SCI.24 This finding is in accordance with our results, which showed the upregulation of Bcl-2 after miR-135a-5p transfection (Figure 3G). In addition, the disruption of the balance between Bax and Bcl-2 was found to contribute to the activation of caspase-3 signaling.51 Caspase-3 was also verified to be a downstream regulator of SP1 by showing that SP1 could activate the caspase-3 promoter.52 Caspase-3 was shown to be the primary activator of apoptotic DNA fragmentation,53 thereby being regarded as an indicator of apoptosis.54 Many previous studies have shown the activation of caspase-3 signaling in the injured spinal cord tissue.49,55,56 In accordance with these results, our study also found the upregulation of cleaved caspase-3 in the miR-135a-5p inhibition-induced apoptosis promotion, as well as the downregulation of cleaved caspase-3 in the miR-135a-5p overexpression-induced apoptosis inhibition. Caspase-3 has also been shown to be downstream of Bcl-2.57 Based on these results, it is reasonable to conclude that miR-135a-5p-SP1 can regulate Bax/Bcl-2/caspase-3 signaling to modulate neuronal apoptosis.
Apart from SP1, miR-135a-5p was also found to be involved in modulating axon regeneration by targeting ROCK1/2. A negative relationship between miR-135a-5p and ROCK was also shown in the present study, using gene transfection assays and animal experiments. The gene transfection assays showed that the expression of ROCK could be increased by inhibiting miR-135a-5p, and vice versa. ROCK expression levels were decreased by transfection of the miR-135a-5p mimic (Figure 4E). Animal experiments also showed that ROCK expression was downregulated after the treatment of miR-135a-5p when compared with the control group and miR-NC group (Figures 7A and 7C). The negative correlation between miR-135a-5p and ROCK1/2 levels may be explained by the fact that miR-135a-5p binds to the 3′ UTR of its target genes, ROCK1 and ROCK2, thereby negatively regulating their expression (Figures 5B and 5C). More importantly, the present study found that the downregulation of ROCK1/2 induced by miR-135a-5p transfection was involved in promoting axon regeneration (Figures 4A, 4B, and 4E). In contrast, rescue experiments showed that silencing ROCK could reverse the inhibition of axon growth induced by the miR-135a-5p inhibitor (Figure 4A, 4B, and 4G). Similar to our results, previous studies have also shown that inhibitors of ROCK can improve axon regeneration,18,19,27 indicating that ROCK inhibition may be a promising therapeutic strategy for SCI.
Axon regeneration induced by the miR-135a-5p-ROCK axis through AKT/GSK3β signaling by activating AKT and inactivating GSK3β (Figure 4E). A previous study has shown that an increase in ROCK activity activates PTEN to downregulate AKT activity,58 which may result in opposing levels of phosphorylated AKT. Activation of the AKT pathway has been shown to regulate neuron regrowth by promoting the sprouting and elongation of axons. In addition, GSK3β is negatively regulated by upstream signaling molecules, including AKT.59 AKT-dependent phosphorylation on Ser9 is a key mechanism that inactivates GSK3β,60 and its inactivation can promote significant axon regeneration.61 Similar to multiple previous studies, our findings show that inhibiting GSK3β activation promoted axon regrowth and thus benefited functional recovery following SCI. Our study also showed opposing trends between AKT and GSK3β activation levels due to the fact that AKT activation inhibits the activity of GSK3β.62 Based on these findings, it is reasonable to conclude that the miR-135a-5p-ROCK axis regulates downstream AKT/GSK3β signaling to modulate axon regeneration.
Considering the strengths of the methodology of the present study, the bioinformatics techniques were first used to identify the critical role of miR-135a-5p in the pathophysiology of SCI. Additionally, various biomolecular experiments were performed to investigate the mechanisms whereby miR-135a-5p affects the functional recovery of SCI, by identifying its target genes and signaling pathways. Another major strength of this study is that it investigated the underlying mechanisms of miR-135a-5p in detail by performing both in vitro and in vivo experiments. However, the limitations of this study should also be acknowledged. The present study only focused on the roles of the miR-135a-5p-SP1-Bax/Bcl-2/caspase-3 and miR-135a-5p-ROCK-AKT/GSK3β axes in regulating neuronal apoptosis and axon regeneration. However, the presence of detailed crosstalk also needs to be explored. Our results suggested that the miR-135a-5p-ROCK-AKT/GSK3β and miR-135a-5p-SP1-Bax/Bcl-2/caspase-3 axes can be regarded as potential therapeutic targets for promoting effective therapy for SCI patients. Moreover, these two signaling axes could be used to genetically manipulate cells and thus benefit the clinical translation of stem cell therapy in SCI.
Materials and Methods
Bioinformatics and Data Mining
To identify the miRNAs that play critical roles in SCI, the datasets investigating the expression profiles of miRNAs were searched in the database: Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information (NCBI).63 The searching criteria were based on the following: t (1) the dataset should be aimed at investigating the expression profiles of miRNAs that were involved in rat SCI; (2) the study design of the dataset should be established by setting the sham group (laminectomy with no spinal cord contusion) and the injury group (laminectomy with spinal cord contusion); and (3) each group should have at least three repeated samples with statistical significance. Based on these criteria, the GEO: GSE19890 dataset was selected to be analyzed in the present study. This dataset investigated multiple time points following SCI, including at the first, third, and seventh day post-injury. The study design of this dataset was established by setting the sham group and injury group at the first, third, and seventh day post-SCI. Finally, five samples in the sham group at the third day (GEO: GSM497216, GSM497217, GSM497218, GSM497219, and GSM497220) and five samples in the injury group at the third day (GEO: GSM497211, GSM497212, GSM497213, GSM497214, and GSM497215) were analyzed to identify the miRNAs that were aberrantly expressed during the SCI pathophysiological process.
After the dataset was selected, the differential expression analysis (DEA) was performed by using the limma package of the R program.64 The miRNAs with p value <0.05 and |log fold change [FC]| >1 were defined as DE miRNAs. The heatmap was plotted based on the expression level of DE miRNAs.
To predict the neurological recovery-related miRNAs, the experimentally-verified miRNAs related to neurological recovery were extracted by the specialized literature mining method. Literature mining is a specialized data mining method that is used to systematically extract information from the text, such as abstracts, or full-text versions of scientific literature.32, 33, 34 In addition, the expression levels of these miRNAs following SCI were obtained from the GEO: GSE19890 dataset. The correlation between DE miRNAs identified in the GEO: GSE19890 dataset and neurological recovery-related miRNAs verified by scientific literature was analyzed based on the PCC analysis. The DE miRNAs with a correlation >0.93 were regarded as DE miRNAs highly correlated with neurological recovery. Using the Jaccard index formula as below, the functional similarity analysis was performed by comparing the functional similarity between literature-verified neurological recovery-related miRNAs (obtained by literature mining) and DE miRNAs highly correlated to neurological recovery (obtained by PCC analysis).
where JI (M, N) indicates the Jaccard index between miRNA, named as M (miRNA-M), and miRNA named as N (miRNA-N); |M∩N| indicates the gene targets, enriched biological processes (BPs), or pathways that are shared between miRNA-M and miRNA-N; in other words, the intersection between two categories; and |M∪N| indicates all gene targets, enriched BPs, or pathways of miR-M and miR-N; in other words, the union between two categories. By performing the functional similarity analysis based on the above formula, miR-135a-5p was found to play a pivotal role in the functional recovery of SCI.
To explore the functions of miR-135a-5p, a functional enrichment analysis (FEA) was performed to investigate the functional terms of its target genes. The target genes of miR-135a-5p were extracted from three databases (Database: miRDB Version 6.0 (http://www.mirdb.org/); Database: miRWalk (http://mirwalk.umm.uni-heidelberg.de/); Database: TargetScan v7.1 (http://www.targetscan.org/vert_71/)),15,65,66 and the union of these three groups of target genes was obtained. The FEA analysis was conducted by using the cluster Profiler package of the R program.67 The functional terms of the target genes were explored by investigating their enriched Gene Ontology (GO) terms, especially BPs, as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The GO terms and KEGG pathways with a p value <0.05 were regarded as significant functional GO. If the number of enriched BPs and pathways was greater than 30, only the top 30 with the highest p value were chosen to be visualized in the bar plot. If the number of enriched BPs and pathways was lower than 30, all of the BPs and pathways were visualized in the bar plot.
PC12 Cell Culture
PC12 cells (Chinese Academy of Sciences, Shanghai, China) were cultured at 37°C in a 5% CO2 environment in RPMI 1640 medium (Gibco, USA) supplemented with 10% heat-inactivated horse serum, 5% fetal bovine serum (FBS) (Gibco, USA), and 100 U/mL penicillin/streptomycin (Gibco, USA). The medium was refreshed every 2 days.
Prior to H2O2 treatment, PC12 cells were seeded in multi-well plates at a density of 5 × 105 cells/well for 24 h. In the treatment group, H2O2 at the concentrations of 25, 50, 100, 200, and 400 μM were utilized to stimulate PC12 cells for 24 h. In the control group, cells were cultured with the medium without H2O2 stimulation.
To induce the neuronal differentiation of PC12 cells, cells were first plated onto culture plates at a density of 5 × 103 cells/well in RPMI 1640 medium with 1% FBS. After culturing for 24 h, a concentrated stock of nerve growth factor (NGF) (human recombinant NGF, Sigma, St. Louis, MO, USA) was then added into culture medium with the final concentration of 100 ng/mL.
Transfection of miRNA Oligonucleotides
The transfection of miRNA oligonucleotides was performed as previously described.68 Briefly, PC12 cells were incubated in serum-free Opti-MEM and then transfected with a miR-135a-5p mimic, a mimic NC, a miR-135a-5p inhibitor, an inhibitor NC, siSP1, siROCK1, siROCK2, or siRNA-NC (RiboBio, Guangzhou, China), at a final concentration of 100 nM, using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) for 6 h.
Apoptosis Assay
Apoptosis was evaluated using an annexin V-FITC/propidium iodide (PI) apoptosis detection kit (Beijing Biosea Biotechnology, Beijing, China). Treated or transfected cells were rinsed twice with PBS. These cells were then stained with 5 μL of annexin V-fluorescein isothiocyanate (FITC) for 15 min at room temperature in the dark. Thereafter, 5 μL of PI was added to the cell suspension. Flow cytometry analysis was performed using a FACScan instrument (Beckman Coulter, Fullerton, CA, USA).
Cell Viability Assay
miRNA-transfected PC12 cells were seeded in 96-well plates at 5 × 103 cells/well, and then treated with H2O2 for 24 h. Cells were then cultured in fresh medium for 24 h at 37°C, and 10 μL of CCK-8 solution (Dojindo Molecular Technologies, Gaithersburg, MD, USA) was added to the culture medium. The cultures were incubated for 1–4 h at 37°C under humidified conditions of 95% air and 5% CO2. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Measurement of Neurite Outgrowth
Cells were visualized under a microscope. Cells extending at least one neurite longer than the cell body diameter were determined as positive neurite-bearing cells, as previously described.68,69 The average neurite length was measured for all neurite-bearing cells in a field by tracing the longest neurite length per effectively transfected cell, using ImageJ software (RRID: SCR_003070). Neurite outgrowth was measured for an average of 100 effectively transfected cells per experiment.
Animal Experiments
Adult female Sprague-Dawley rats weighing 200–250 g were purchased from the Vital River Laboratory Animal Technology (Beijing, China). The experimental animal protocols, including care, breeding, and operative procedures, were approved by the Animal Care and Use Committee of South China Agricultural University (2019-D096).
Establishment of SCI Model
Rats were anesthetized via an intraperitoneal injection of 2% sodium pentobarbital (0.25 mL/100 g).70 Following the dissection of the paraspinal muscles, a laminectomy from T10 was performed. A New York University Impactor was used as previously described.71 SCI was induced in rats using a 10-g weight drop from a height of 50 mm. Rats in the sham group received only a T10 laminectomy without the weight-drop injury. Finally, the incision was closed in layers. After surgery, all animals received penicillin and analgesics for 3 consecutive days, and the bladders were manually voided twice daily. The animals received treatments by intrathecal injection after SCI.
Experimental Design
Twelve rats were randomly divided into four groups: sham group and SCI group (1 day, 2 days, and 3 days after SCI) (n = 3 per group). The parenchymal tissue at the surgery site was extracted for western blotting and qRT-PCR analysis, as previously described. The remaining 48 rats were randomly divided into the following four groups, and testing was performed by blinded observers: (1) sham group (the rats were subjected to sham operation), (2) control group (the rats were subjected to SCI and were treated with PBS), (3) agomir-NC group (the rats were subjected to SCI and treated with agomir-NC [RiboBio, Guangzhou, China]), and (4) agomir-135a-5p group (the rats were subjected to SCI and treated with 25 μL of 10 nmol agomir-135a-5p [RiboBio, Guangzhou, China]) (n = 12 per group). The damaged spinal cords were collected 7 days after intrathecal injection for further western blotting and immunofluorescence analyses.
Evaluation of Functional Recovery
The BBB scale, ranging from 0 to 21, was used to assess motor function recovery. A score of 0 indicated complete paralysis of the hind limb, and a score of 21 indicated normal function. Rats were independently observed in an open-field environment at different time periods (1, 3, 7, 10, 14, 17, 21, 24, and 28 days after SCI) by two experienced, blinded investigators. The mean values based on the BBB scale values from the two assessors were recorded for each group.
qRT-PCR assay
Total RNA was extracted by using an RNeasy mini kit (QIAGEN, Valencia, CA, USA), and RNA concentration and quality were determined by measuring absorbance at 260 nm (A260) and calculating the A260/A280 ratio, respectively. miR-135a-5p levels were quantified by qRT-PCR, with U6 small nuclear RNA as an internal reference for normalization. Bulge-Loop miRNA qRT-PCR primer sets (one RT primer and a pair of qPCR primers for each set) specific for miR-135a-5p were designed by Ribobio. The sequences of the miR-135a-5p RT primer, miR reverse primer, and miR-135a-5p forward primer are proprietary. The relative expression levels normalized to U6 levels were calculated using the 2−ΔΔCt method. For mRNA analysis, total RNA was reverse transcribed using RT (Takara Bio, Kusatsu, Japan), according to the manufacturer’s instructions. qRT-PCR was performed using multiple kits (SYBR Green I master; Roche, Basel, Switzerland) and a LightCycler 480 Instrument (Roche). Each gene was normalized to the expression level of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Western Blotting Analysis
Total protein was extracted from tissues or cells using radioimmunoprecipitation assay cell lysis buffer, containing phenylmethanesulfonyl fluoride and transferred onto polyvinylidene fluoride membranes using a wet transfer method. The membranes were then incubated with diluted primary antibodies (Abcam, Cambridge, UK). These included antibodies targeting Sp1 (ab227383, 1:2,000), Bax (ab182733, 1:2,000), Bcl-2 (ab196495, 1:1,000), pro-caspase-3 (ab32150, 1:1,000), cleaved caspase-3 (ab49822, 1:500), ROCK1 (ab45171, 1:5,000), ROCK2 (ab125025, 1:10,000), GSK3β (ab93926, 1:1,000), phosphorylated GSK3β (p-GSK3β, Ser9, ab131097, 1:1,000), NF-200 (ab134306, 1:500), GAP43 (ab75810, 1:1,000), GAPDH (ab245355, 1:10,000), AKT (pan-AKT, ab179463, 1:1,000), phosphorylated AKT (p-AKT, Thr308, ab38449, 1:500), and phosphorylated AKT (p-AKT, Ser473, 4060s, 1:500, Cell Signaling Technology, San Diego, CA, USA). All of these antibodies were incubated overnight at 4°C. After incubation with primary antibody, the membranes were incubated with a horseradish peroxidase-tagged secondary goat anti-rabbit immunoglobulin G (IgG) antibody (ab97051, 1:2,000, Abcam) for 1 h. Images were captured using a Bio-Rad image analysis system. Relative protein expression levels were expressed as the ratio of the target band to the loading control band.
Luciferase Reporter Assay
A luciferase reporter system was used to further clarify the interaction between miR-135a-5p and its targets (SP1, ROCK1, and ROCK2). First, WT plasmids pGL3-SP1-3ʹ UTRs (SP1 3ʹ UTR WT), pGL3-ROCK1-3ʹ UTRs (ROCK1 3ʹ UTR WT), pGL3-ROCK2-3ʹ UTRs (ROCK2 3ʹ UTR WT), and MUT plasmids, including pGL3-SP1-3ʹ UTR (SP1 3ʹ UTR MUT), pGL3-ROCK1-3ʹ UTR (ROCK1 3ʹ UTR MUT), and pGL3-ROCK2-3ʹ UTR (ROCK2 3ʹ UTR MUT), and their null plasmids were created. HEK293 cells were added to 12-well plates 24 h before transfection (1 × 105 cells/well), and 200 ng of recombinant plasmid was co-transfected into the cells with 100 nM miR-135a-5p mimic or the negative control. Simultaneously, pRL-TK, which expresses Renilla luciferase (RL), was transfected as an internal reference. After 24 h, the intensities of the fluorescence generated by firefly luciferase and RL were measured, and changes in the ratio of the two fluorescence intensities were used to verify the interaction between miR-135a-5p and its targets (SP1, ROCK1, and ROCK2 3ʹ UTRs). Each sample was assayed in three duplicate wells.
Immunohistochemistry
Animals were anesthetized terminally with an overdose of isoflurane inhalation on day 7 after intrathecal transplantation. The spinal cords were embedded in optimal cutting temperature compound. T9–T11 spinal cord segments containing the lesion epicenter were collected for histological evaluation. Briefly, longitudinal sections were deparaffinized with xylene, hydrated in a graded alcohol series, and boiled in citrate buffer (pH 6.0) twice for 5 min each. Subsequently, the sections were cooled and incubated in 3% H2O2 for 15 min at room temperature to inactivate endogenous peroxidases. The slides were then blocked with 10% FBS for 10 min and incubated with rabbit anti-GAP43 or rabbit anti-NF (Abcam) primary antibodies overnight at 4°C. After washing with Tris-buffered saline (TBS), the sections were incubated with fluorescently labeled secondary antibodies (Abcam). Finally, the sections were stained with DAPI and visualized under a confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany).
Statistical Analysis
Data are presented as the mean ± standard deviation and were analyzed using a Student’s t test or one-way analysis of variance. All statistical analyses were performed using SPSS 22.0 (IBM, Armonk, NY, USA) or Prism 8 (GraphPad, San Diego, CA, USA) software. A p value <0.05 indicated statistical significance.
Author Contributions
N.W., Y.Y., and M.P. designed the study and experimental work. C.D. and Y.C. participated in sample collection and data analysis. S.L. carried out bioinformatics analyses. Z.T. and F.F. produced the initial draft of the manuscript. Y.W. and Z.C. contributed to drafting the manuscript. B.L. and L.R. conceived the study design and participated in the analysis and interpretation of the data. All of the authors have approved the final drafting and revised all versions of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2017YFA0105403); the Key Research and Development Program of Guangdong Province (2019B020236002); the Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201006); the National Natural Science Foundation of China (31470949 and 81772349); the Guangdong Basic and Applied Basic Research Foundation (2020A1515010171 and 2019A1515110814); the Guangzhou Health Care Cooperative Innovation Major Project (201704020221); the Guangzhou Science and Technology Project (201707010115 and 201704020049); the Guangdong Natural Science Foundation (2017A030313594); and by the Medical Scientific Research Foundation of Guangdong Province (A2018547).
References
- 1.Hong J.Y., Lee S.H., Lee S.C., Kim J.W., Kim K.P., Kim S.M., Tapia N., Lim K.T., Kim J., Ahn H.S. Therapeutic potential of induced neural stem cells for spinal cord injury. J. Biol. Chem. 2014;289:32512–32525. doi: 10.1074/jbc.M114.588871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khayrullina G., Bermudez S., Byrnes K.R. Inhibition of NOX2 reduces locomotor impairment, inflammation, and oxidative stress after spinal cord injury. J. Neuroinflammation. 2015;12:172. doi: 10.1186/s12974-015-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R., Yin F., Guo Y.Y., Zhao K.C., Ruan Q., Qi Y.M. Knockdown of ANRIL aggravates H2O2-induced injury in PC-12 cells by targeting microRNA-125a. Biomed. Pharmacother. 2017;92:952–961. doi: 10.1016/j.biopha.2017.05.122. [DOI] [PubMed] [Google Scholar]
- 4.Özdemir Ü.S., Nazıroğlu M., Şenol N., Ghazizadeh V. Hypericum perforatum attenuates spinal cord injury-induced oxidative stress and apoptosis in the dorsal root ganglion of rats: involvement of TRPM2 and TRPV1 channels. Mol. Neurobiol. 2016;53:3540–3551. doi: 10.1007/s12035-015-9292-1. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Chen X., Huang X., Qin C., Fang Y., Liu Y., Zhang G., Pan D., Wang W., Xie M. Soluble epoxide hydrolase inhibition provides multi-target therapeutic effects in rats after spinal cord injury. Mol. Neurobiol. 2016;53:1565–1578. doi: 10.1007/s12035-015-9118-1. [DOI] [PubMed] [Google Scholar]
- 6.Wee H.Y., Lim S.W., Chio C.C., Niu K.C., Wang C.C., Kuo J.R. Hyperbaric oxygen effects on neuronal apoptosis associations in a traumatic brain injury rat model. J. Surg. Res. 2015;197:382–389. doi: 10.1016/j.jss.2015.04.052. [DOI] [PubMed] [Google Scholar]
- 7.Shin J.E., Cho Y. Epigenetic regulation of axon regeneration after neural injury. Mol. Cells. 2017;40:10–16. doi: 10.14348/molcells.2017.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahar M., Cavalli V. Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 2018;19:323–337. doi: 10.1038/s41583-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li P., Teng Z.-Q., Liu C.-M. Extrinsic and intrinsic regulation of axon regeneration by microRNAs after spinal cord injury. Neural Plast. 2016;2016:1279051. doi: 10.1155/2016/1279051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan H., Hong P., Jiang M., Li H. MicroRNAs as potential therapeutics for treating spinal cord injury. Neural Regen. Res. 2012;7:1352–1359. doi: 10.3969/j.issn.1673-5374.2012.17.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou S., Ding F., Gu X. Non-coding RNAs as emerging regulators of neural injury responses and regeneration. Neurosci. Bull. 2016;32:253–264. doi: 10.1007/s12264-016-0028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Battum E.Y., Verhagen M.G., Vangoor V.R., Fujita Y., Derijck A.A.H.A., O’Duibhir E., Giuliani G., de Gunst T., Adolfs Y., Lelieveld D. An image-based miRNA screen identifies miRNA-135s as regulators of CNS axon growth and regeneration by targeting Krüppel-like factor 4. J. Neurosci. 2018;38:613–630. doi: 10.1523/JNEUROSCI.0662-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Liao S., Quan H., Lin Y., Li J., Yang Q. Involvement of microRNA-135a-5p in the protective effects of hydrogen sulfide against Parkinson’s disease. Cell. Physiol. Biochem. 2016;40:18–26. doi: 10.1159/000452521. [DOI] [PubMed] [Google Scholar]
- 15.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deniaud E., Baguet J., Mathieu A.L., Pagès G., Marvel J., Leverrier Y. Overexpression of Sp1 transcription factor induces apoptosis. Oncogene. 2006;25:7096–7105. doi: 10.1038/sj.onc.1209696. [DOI] [PubMed] [Google Scholar]
- 17.García-Morales V., Rodríguez-Bey G., Gómez-Pérez L., Domínguez-Vías G., González-Forero D., Portillo F., Campos-Caro A., Gento-Caro Á., Issaoui N., Soler R.M. Sp1-regulated expression of p11 contributes to motor neuron degeneration by membrane insertion of TASK1. Nat. Commun. 2019;10:3784. doi: 10.1038/s41467-019-11637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch J.C., Tönges L., Barski E., Michel U., Bähr M., Lingor P. ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS. Cell Death Dis. 2014;5:e1225. doi: 10.1038/cddis.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi A.R., Muke I., Bobylev I., Lehmann H.C. ROCK inhibition improves axonal regeneration in a preclinical model of amyotrophic lateral sclerosis. J. Comp. Neurol. 2019;527:2334–2340. doi: 10.1002/cne.24679. [DOI] [PubMed] [Google Scholar]
- 20.Ryge J., Winther O., Wienecke J., Sandelin A., Westerdahl A.C., Hultborn H., Kiehn O. Transcriptional regulation of gene expression clusters in motor neurons following spinal cord injury. BMC Genomics. 2010;11:365. doi: 10.1186/1471-2164-11-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torabi B., Flashner S., Beishline K., Sowash A., Donovan K., Bassett G., Azizkhan-Clifford J. Caspase cleavage of transcription factor Sp1 enhances apoptosis. Apoptosis. 2018;23:65–78. doi: 10.1007/s10495-017-1437-4. [DOI] [PubMed] [Google Scholar]
- 22.Uchida A., Oh-hashi K., Kiuchi K., Hirata Y. Manganese regulates caspase-3 gene promoter activity by inducing Sp1 phosphorylation in PC12 cells. Toxicology. 2012;302:292–298. doi: 10.1016/j.tox.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Kotipatruni R.R., Dasari V.R., Veeravalli K.K., Dinh D.H., Fassett D., Rao J.S. p53- and Bax-mediated apoptosis in injured rat spinal cord. Neurochem. Res. 2011;36:2063–2074. doi: 10.1007/s11064-011-0530-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Sun Z.Y., Zhang K.M., Xu G.Q., Li G. Bcl-2 in suppressing neuronal apoptosis after spinal cord injury. World J. Emerg. Med. 2011;2:38–44. [PMC free article] [PubMed] [Google Scholar]
- 25.Shin J.-Y., Kim Y.-I., Cho S.-J., Lee M.K., Kook M.-C., Lee J.H., Lee S.S., Ashktorab H., Smoot D.T., Ryu K.W. MicroRNA 135a suppresses lymph node metastasis through down-regulation of ROCK1 in early gastric cancer. PLoS ONE. 2014;9:e85205. doi: 10.1371/journal.pone.0085205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroiss A., Vincent S., Decaussin-Petrucci M., Meugnier E., Viallet J., Ruffion A., Chalmel F., Samarut J., Allioli N. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene. 2015;34:2846–2855. doi: 10.1038/onc.2014.222. [DOI] [PubMed] [Google Scholar]
- 27.Fujita Y., Yamashita T. Axon growth inhibition by RhoA/ROCK in the central nervous system. Front. Neurosci. 2014;8:338. doi: 10.3389/fnins.2014.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vemula S., Shi J., Hanneman P., Wei L., Kapur R. ROCK1 functions as a suppressor of inflammatory cell migration by regulating PTEN phosphorylation and stability. Blood. 2010;115:1785–1796. doi: 10.1182/blood-2009-08-237222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H., Miao L., Yang L., Liang F., Wang Q., Zhuang P., Sun Y., Hu Y. AKT-dependent and -independent pathways mediate PTEN deletion-induced CNS axon regeneration. Cell Death Dis. 2019;10:203. doi: 10.1038/s41419-018-1289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liz M.A., Mar F.M., Santos T.E., Pimentel H.I., Marques A.M., Morgado M.M., Vieira S., Sousa V.F., Pemble H., Wittmann T. Neuronal deletion of GSK3β increases microtubule speed in the growth cone and enhances axon regeneration via CRMP-2 and independently of MAP1B and CLASP2. BMC Biol. 2014;12:47. doi: 10.1186/1741-7007-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masic I., Milinovic K. On-line biomedical databases-the best source for quick search of the scientific information in the biomedicine. Acta Inform. Med. 2012;20:72–84. doi: 10.5455/aim.2012.20.72-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen L.J., Saric J., Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 33.Feng L., Chiam Y., Lo S.K. 2017. Text-mining techniques and tools for systematic literature reviews: a systematic literature review. In Proceedings of the 24th Asia-Pacific Software Engineering Conference (APSEC) pp. 41–50. [Google Scholar]
- 34.Korhonen A., Séaghdha D.O., Silins I., Sun L., Högberg J., Stenius U. Text mining for literature review and knowledge discovery in cancer risk assessment and research. PLoS ONE. 2012;7:e33427. doi: 10.1371/journal.pone.0033427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li R., Zhao K., Ruan Q., Meng C., Yin F. Bone marrow mesenchymal stem cell-derived exosomal microRNA-124-3p attenuates neurological damage in spinal cord ischemia-reperfusion injury by downregulating Ern1 and promoting M2 macrophage polarization. Arthritis Res. Ther. 2020;22:75. doi: 10.1186/s13075-020-2146-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan S., Wang Y.X., Gong P.H., Meng C.Y. miR-124 inhibits spinal neuronal apoptosis through binding to GCH1. Eur. Rev. Med. Pharmacol. Sci. 2019;23:4564–4574. doi: 10.26355/eurrev_201906_18032. [DOI] [PubMed] [Google Scholar]
- 37.Louw A.M., Kolar M.K., Novikova L.N., Kingham P.J., Wiberg M., Kjems J., Novikov L.N. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomedicine (Lond.) 2016;12:643–653. doi: 10.1016/j.nano.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Wang N., He L., Yang Y., Li S., Chen Y., Tian Z., Ji Y., Wang Y., Pang M., Wang Y. Integrated analysis of competing endogenous RNA (ceRNA) networks in subacute stage of spinal cord injury. Gene. 2020;726:144171. doi: 10.1016/j.gene.2019.144171. [DOI] [PubMed] [Google Scholar]
- 39.Hong E.S., Yao H.H., Min Y.J., Sun J., Zhou X., Zeng X.B., Yu W. The mechanism of electroacupuncture for treating spinal cord injury rats by mediating Rho/Rho-associated kinase signaling pathway. J. Spinal Cord Med. 2019 doi: 10.1080/10790268.2019.1665612. Published online October 9, 2019. 10.1080/10790268.2019.1665612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu N., Shi Y.F., Diao H.Y., Li Y.X., Cui Y., Song X.J., Tian X., Li T.Y., Liu B. MicroRNA-135a regulates apoptosis induced by hydrogen peroxide in rat cardiomyoblast cells. Int. J. Biol. Sci. 2017;13:13–21. doi: 10.7150/ijbs.16769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu H., Lee J., Zaman K., Kubilis J., Ferrante R.J., Ross B.D., Neve R., Ratan R.R. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J. Neurosci. 2003;23:3597–3606. doi: 10.1523/JNEUROSCI.23-09-03597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh S.H., Yang W.B., Gean P.W., Hsu C.Y., Tseng J.T., Su T.P., Chang W.C., Hung J.J. Translational and transcriptional control of Sp1 against ischaemia through a hydrogen peroxide-activated internal ribosomal entry site pathway. Nucleic Acids Res. 2011;39:5412–5423. doi: 10.1093/nar/gkr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J., Kosaras B., Aleyasin H., Han J.A., Park D.S., Ratan R.R., Kowall N.W., Ferrante R.J., Lee S.W., Ryu H. Role of cyclooxygenase-2 induction by transcription factor Sp1 and Sp3 in neuronal oxidative and DNA damage response. FASEB J. 2006;20:2375–2377. doi: 10.1096/fj.06-5957fje. [DOI] [PubMed] [Google Scholar]
- 44.Ryu H., Lee J., Olofsson B.A., Mwidau A., Dedeoglu A., Escudero M., Flemington E., Azizkhan-Clifford J., Ferrante R.J., Ratan R.R. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proc. Natl. Acad. Sci. USA. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pietrzak M., Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol. Chem. 2008;389:383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt T., Körner K., Karsunky H., Korsmeyer S., Müller R., Möröy T. The activity of the murine Bax promoter is regulated by Sp1/3 and E-box binding proteins but not by p53. Cell Death Differ. 1999;6:873–882. doi: 10.1038/sj.cdd.4400562. [DOI] [PubMed] [Google Scholar]
- 47.Pawlowski J., Kraft A.S. Bax-induced apoptotic cell death. Proc. Natl. Acad. Sci. USA. 2000;97:529–531. doi: 10.1073/pnas.97.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin L.J., Liu Z. Injury-induced spinal motor neuron apoptosis is preceded by DNA single-strand breaks and is p53- and Bax-dependent. J. Neurobiol. 2002;50:181–197. doi: 10.1002/neu.10026. [DOI] [PubMed] [Google Scholar]
- 49.Springer J.E., Azbill R.D., Knapp P.E. Activation of the caspase-3 apoptotic cascade in traumatic spinal cord injury. Nat. Med. 1999;5:943–946. doi: 10.1038/11387. [DOI] [PubMed] [Google Scholar]
- 50.Or C.R., Huang C.W., Chang C.C., Lai Y.C., Chen Y.J., Chang C.C. Obatoclax, a pan-BCL-2 inhibitor, downregulates survivin to induce apoptosis in human colorectal carcinoma cells via suppressing WNT/β-catenin signaling. Int. J. Mol. Sci. 2020;21:1773. doi: 10.3390/ijms21051773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang B., Johnson T.S., Thomas G.L., Watson P.F., Wagner B., Furness P.N., El Nahas A.M. A shift in the Bax/Bcl-2 balance may activate caspase-3 and modulate apoptosis in experimental glomerulonephritis. Kidney Int. 2002;62:1301–1313. doi: 10.1111/j.1523-1755.2002.kid587.x. [DOI] [PubMed] [Google Scholar]
- 52.Sudhakar C., Jain N., Swarup G. Sp1-like sequences mediate human caspase-3 promoter activation by p73 and cisplatin. FEBS J. 2008;275:2200–2213. doi: 10.1111/j.1742-4658.2008.06373.x. [DOI] [PubMed] [Google Scholar]
- 53.Wolf B.B., Schuler M., Echeverri F., Green D.R. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J. Biol. Chem. 1999;274:30651–30656. doi: 10.1074/jbc.274.43.30651. [DOI] [PubMed] [Google Scholar]
- 54.BD Biosciences . 2012. Caspase-3 activation-an indicator of apoptosis in image-based assays.https://beta-static.fishersci.com/content/dam/fishersci/en_US/documents/programs/scientific/technical-documents/application-notes/caspase-3-activiation-indicator-apoptosis-application-notes.pdf [Google Scholar]
- 55.Citron B.A., Arnold P.M., Sebastian C., Qin F., Malladi S., Ameenuddin S., Landis M.E., Festoff B.W. Rapid upregulation of caspase-3 in rat spinal cord after injury: mRNA, protein, and cellular localization correlates with apoptotic cell death. Exp. Neurol. 2000;166:213–226. doi: 10.1006/exnr.2000.7523. [DOI] [PubMed] [Google Scholar]
- 56.Ozawa H., Keane R.W., Marcillo A.E., Diaz P.H., Dietrich W.D. Therapeutic strategies targeting caspase inhibition following spinal cord injury in rats. Exp. Neurol. 2002;177:306–313. doi: 10.1006/exnr.2002.7998. [DOI] [PubMed] [Google Scholar]
- 57.Hua F., Cornejo M.G., Cardone M.H., Stokes C.L., Lauffenburger D.A. Effects of Bcl-2 levels on Fas signaling-induced caspase-3 activation: molecular genetic tests of computational model predictions. J. Immunol. 2005;175:985–995. doi: 10.4049/jimmunol.175.2.985. [DOI] [PubMed] [Google Scholar]
- 58.Yang S., Kim H.M. The RhoA-ROCK-PTEN pathway as a molecular switch for anchorage dependent cell behavior. Biomaterials. 2012;33:2902–2915. doi: 10.1016/j.biomaterials.2011.12.051. [DOI] [PubMed] [Google Scholar]
- 59.Luo L., Chen J., Su D., Chen M., Luo B., Pi R., Wang L., Shen W., Wang R. L-F001, a multifunction ROCK inhibitor prevents 6-OHDA induced cell death through activating Akt/GSK-3beta and Nrf2/HO-1 signaling pathway in PC12 cells and attenuates MPTP-induced dopamine neuron toxicity in mice. Neurochem. Res. 2017;42:615–624. doi: 10.1007/s11064-016-2117-4. [DOI] [PubMed] [Google Scholar]
- 60.Ambacher K.K., Pitzul K.B., Karajgikar M., Hamilton A., Ferguson S.S., Cregan S.P. The JNK- and AKT/GSK3β- signaling pathways converge to regulate Puma induction and neuronal apoptosis induced by trophic factor deprivation. PLoS ONE. 2012;7:e46885. doi: 10.1371/journal.pone.0046885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J., Zhou Q., Liu C., Liu C., Saijilafu Glycogen synthase kinase 3: a crucial regulator of axotomy-induced axon regeneration. Neural Regen. Res. 2020;15:859–860. doi: 10.4103/1673-5374.268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saraceno G.E., Bellini M.J., Garcia-Segura L.M., Capani F. Estradiol activates PI3K/Akt/GSK3 pathway under chronic neurodegenerative conditions triggered by perinatal asphyxia. Front. Pharmacol. 2018;9:335. doi: 10.3389/fphar.2018.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clough E., Barrett T. The Gene Expression Omnibus database. Methods Mol. Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 67.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin L.F., Chiu S.P., Wu M.J., Chen P.Y., Yen J.H. Luteolin induces microRNA-132 expression and modulates neurite outgrowth in PC12 cells. PLoS ONE. 2012;7:e43304. doi: 10.1371/journal.pone.0043304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hashimoto K., Ishima T. A novel target of action of minocycline in NGF-induced neurite outgrowth in PC12 cells: translation initiation factor eIF4AI. PLoS ONE. 2010;5:e15430. doi: 10.1371/journal.pone.0015430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishio T. Axonal regeneration and neural network reconstruction in mammalian CNS. J. Neurol. 2009;256(Suppl 3):306–309. doi: 10.1007/s00415-009-5244-x. [DOI] [PubMed] [Google Scholar]
- 71.Khuyagbaatar B., Kim K., Kim Y.H. Conversion equation between the drop height in the New York University impactor and the impact force in the Infinite Horizon impactor in the contusion spinal cord injury model. J. Neurotrauma. 2015;32:1987–1993. doi: 10.1089/neu.2015.3875. [DOI] [PubMed] [Google Scholar]