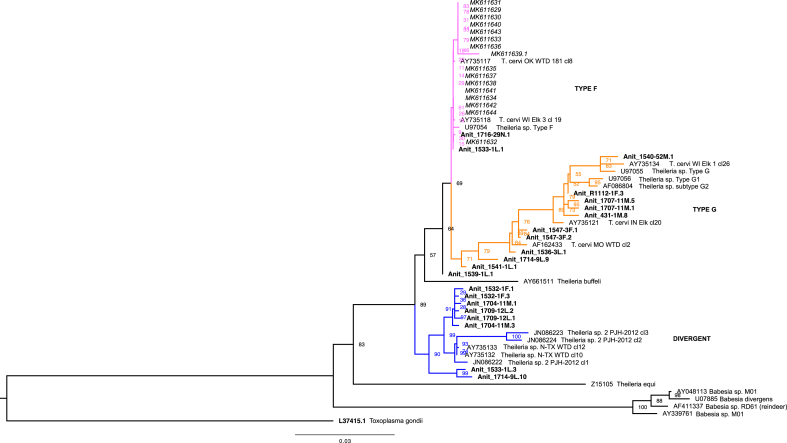
Fig. 3.
Phylogentic analysis of Theileria sp. fragments from Anocenter nitens ticks. Representative Type F, Type G, and ‘divergent’ Theileria sp. sequences were identified from individual A. nitens ticks collected from white-tailed deer and a single nilgai host (bold labels). A maximum-likelihood tree was constructed using Toxoplasma gondii as the outgroup, as it is from a different axpicomplexan class than Theileria. Branch support was assessed with 10,000 replicates of UFBoot bootstrap replication, and bootstrap percentages are indicated at each branch point in the tree. GenBank accession numbers and annotated identification for sequences used in the comparative analysis are indicated on the branch labels. Accession numbers in italics are those T. cervi sequences from white-tailed deer on the East Foundation's San Antonio Viejo Ranch in Starr and Jim Hogg Counties, Texas (Yu et al., 2020).