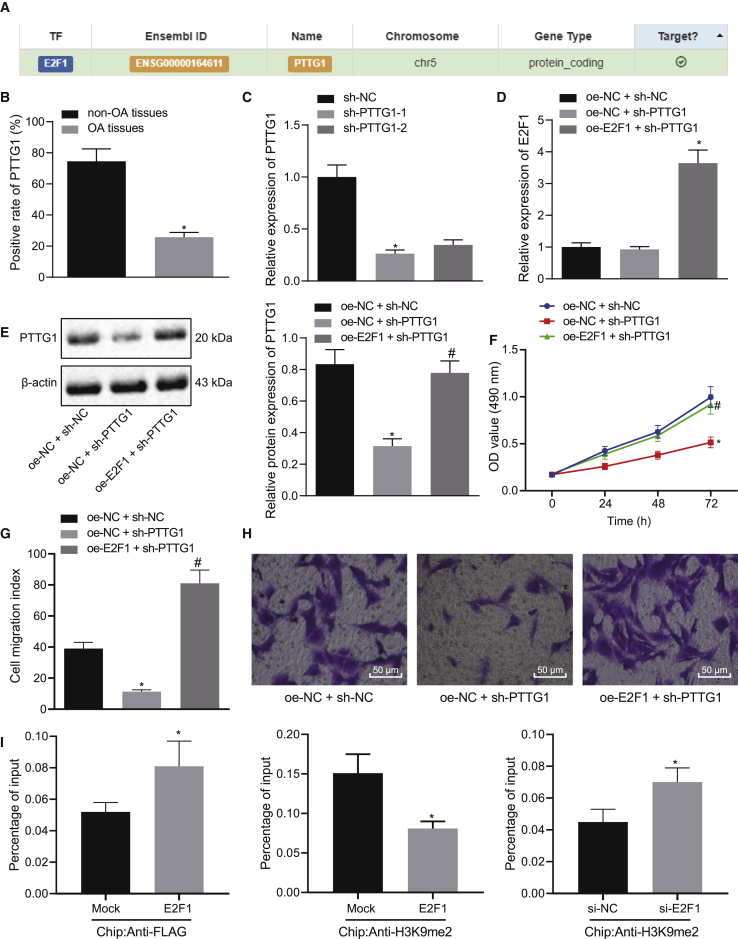
Figure 4.
E2F1 Elevates Transcriptional Activation of the PTTG1 Gene and Augments Chondrocyte Proliferation In Vitro
(A) Targeting relationship between E2F1 and PTTG1 retrieved in the hTFtarget website. (B) Immunohistochemistry of PTTG1 protein in articular cartilage tissues of OA (N = 54) and non-OA (N = 36) subjects. ∗p < 0.05 articular cartilage tissues of non-OA subjects. (C) Transfection efficiency of the sh-PTTG1 determined by qRT-PCR in chondrocytes. ∗p < 0.05 compared with cells treated with sh-NC. (D) mRNA expression of E2F1 determined by qRT-PCR in chondrocytes in response to transfection with sh-PTTG1 or combined with oe-E2F1. (E) Representative western blots of PTTG1 protein and its quantitation in chondrocytes in response to transfection with sh-PTTG1 or combined with oe-E2F1. ∗p < 0.05 compared with chondrocytes transfected with oe-NC + sh-NC; #p < 0.05 compared with chondrocytes transfected with oe-E2F1 + sh-NC. (F) Chondrocyte proliferation measured by the CCK-8 assay in response to transfection with sh-PTTG1 or combined with oe-E2F1. ∗p < 0.05 compared with chondrocytes transfected with oe-NC + sh-NC; #p < 0.05 compared with chondrocytes transfected with oe-E2F1 + sh-NC. (G) The number of migrated chondrocytes measured by the Transwell assay in response to sh-PTTG1 or combined with oe-E2F1. ∗p < 0.05 compared with chondrocytes transfected with oe-NC + sh-NC; #p < 0.05 compared with chondrocytes transfected with oe-E2F1 + sh-NC. (H) Representative images of migrating chondrocytes (×200). (I) The promoter region of the PTTG1 gene analyzed by microarray. The top indicates a schematic of the PTTG1 promoter region. The location of amplified fragments by qRT-PCR was based on the number of nucleotides and related to the transcription start site (TSS) (amplicon). Primer pairs are used for qRT-PCR analysis (detailed information can be seen in Materials and Methods). The intermediate, crosslinked, and sheared chromatin was immunoprecipitated with anti-FLAG antibody (left) and anti-H3K9me2 antibody (right). When E2F1 was increased, H3K9me2 was decreased, with the signal displayed as the percentage of input chromatin (bottom). The average standard deviation of three independent experiments was 6. The p value was calculated by independent sample t test (∗p < 0.05). At the bottom, we employed the anti-H3K9me2 antibody to the ChIP experiment and treated the chondrocytes with si-NC and si-E2F1. The average standard deviation of three independent experiments was 6. The p value was calculated by independent sample t test (∗p < 0.05). Data are shown as mean ± standard deviation of three technical replicates. Data comparison among multiple groups was performed using one-way ANOVA with Tukey’s post hoc test. Data comparison between groups at different time points was performed using repeated-measures ANOVA with Bonferroni correction.