Figure 3.
RNA Sequence and Structure Encode Interactions with N-Protein to Specify Condensation or Dissolution
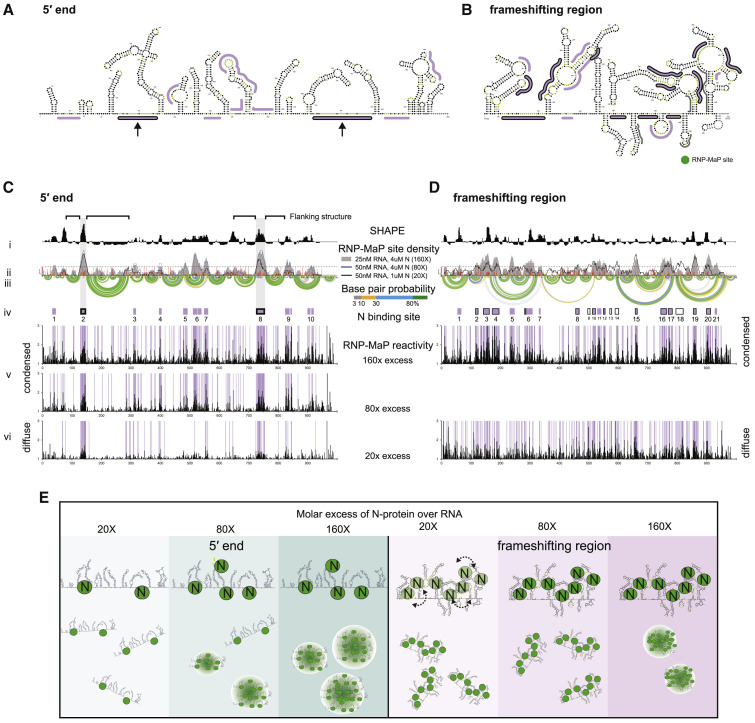
(A and B) SHAPE-Map secondary structure models for the 5′ end (A) or frameshifting region (B). RNP-MaP N-protein binding sites are marked by lines. Two principal binding sites on the 5′ end RNA, both flanked by strong RNA structures, are emphasized with arrows.
(C and D) 5′ end (C) and frameshifting region (D) display condition-specific RNP-MAP reactivity in condensed (80×, 160×) and diffuse (20×) conditions. x axis is the position in nucleotides, y axis is the reactivity (SHAPE or RNP-Map). (i) Windowed (15 nt windows) median SHAPE reactivity (black). (ii) RNP-MaP site density (sites per 15 nt windows); individual nt SHAPE reactivities in colored histograms. (iii) Arcs indicate base pair probabilities (from SHAPE). (iv) N-protein binding sites (boxes: purple, at 160×; with black border, 20×, purple with black border, in 160× and 20×). (v–vi) Raw RNP-MaP reactivity (black) in all conditions. Purple shading highlights RNP-MaP sites.
(E) Model for LLPS. Left panel: 5′ end LLPS coincides with an increase in valency with specific N-protein binding sites. Right panel: frameshifting region RNA has many binding sites (dashed arrows: ensembles of binding sites at lower N concentrations) that enrich N-protein and prevent condensate formation, unless excess N-protein is present to drive LLPS via protein-protein interaction.