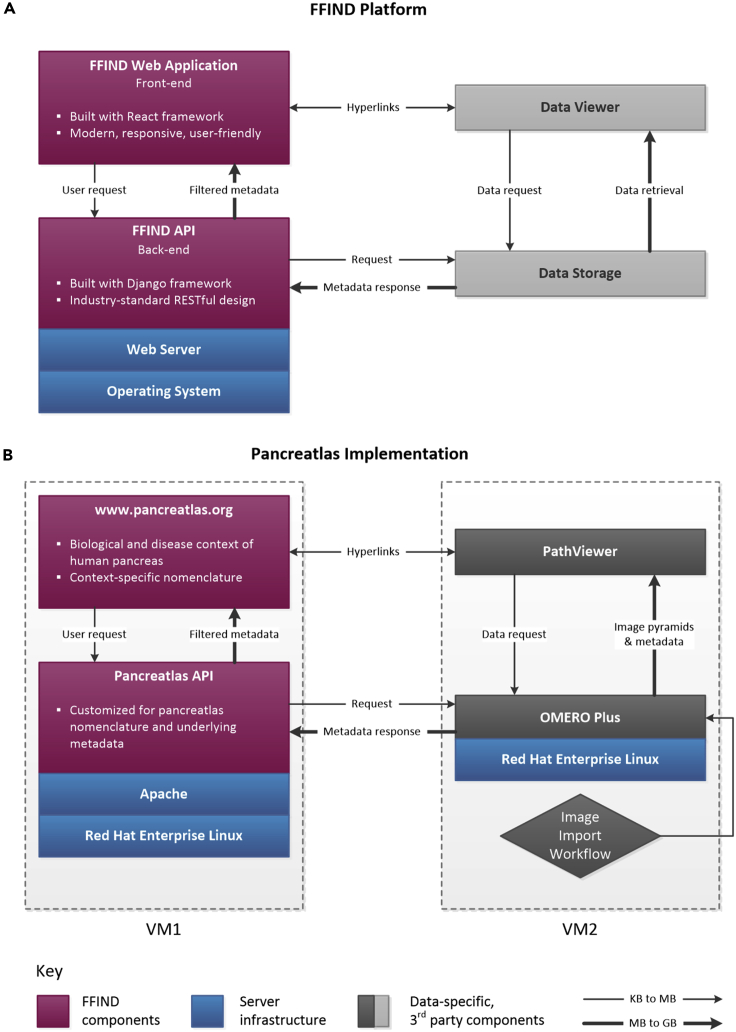
Figure 1.
FFIND Platform and Its Pancreatlas Implementation
Schematic showing general connectivity and data flow across (A) FFIND platform and (B) Pancreatlas implementation. In both panels, unique components are depicted in magenta, shown in relationship to server infrastructure (blue) and third-party components (gray). The thin arrows represent small data requests and large arrows represent larger data responses.
(A) The intuitive web application of FFIND allows end users (researchers, clinicians, biologists) to seamlessly browse modular datasets annotated with field-specific metadata, and it can also launch existing data visualization clients to provide a cohesive browsing experience. The FFIND application programming interface (API) connects the web application to an underlying data server or storage component, which can be configured according to project needs.
(B) To create Pancreatlas, the OMERO Plus server was installed on a virtual machine (VM2) for image management. Data and associated metadata were loaded directly from file stores and proprietary imaging servers (not pictured here), and the web application was built to retrieve and display images in PathViewer, the web client associated with the OMERO Plus server.
See also Table S1.