Figure 1.
Tumor Genomic Features Associated with Clinical Benefit
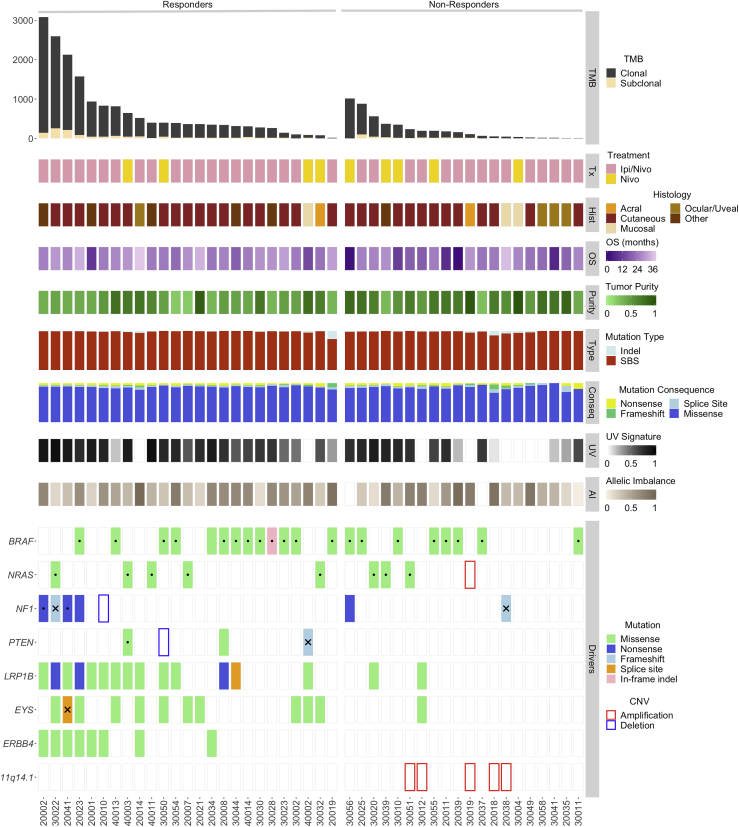
Tumors of responding patients had a higher total and clonal TMB compared to non-responders (FDR-adjusted p = 0.0048 and p = 0.0037, respectively). Overall, a higher number of single-base substitutions and indels were found in tumors of responders, which was largely driven by their higher TMB. A UV-related mutational signature was found to be enriched in tumors of responders for all patients and patients in the ipilimumab/nivolumab group (FDR-adjusted p = 0.03 and p = 0.0096, respectively). Following an exome-wide unbiased approach, we investigated potential differential abundance of sequence alterations in tumors of responding patients. LRP1B and EYS mutations appeared to accumulate in tumors of responding patients (FDR p = 0.058 for both genes and TMB-adjusted p = 0.036 and p = 0.025 for LRP1B and EYS, respectively), most likely due to the expected larger number of passenger mutations in larger DNA regions. There was a non-significant trend in enrichment of ERBB4 mutations in tumors of responders, likely reflecting TMB-high tumors (TMB-adjusted p = 0.133). There were no differences in the abundance of BRAF and NF1 mutations between tumors of responders and non-responders. The AAMDC, CLNS1A, INTS4, KCTD14, NDUFC2, NDUFC2-KCTD14, RSF1, and THRSP loci on chromosome 11q14.1 were found to be co-amplified in five tumors of non-responders (FDR-adjusted p = 0.094). Genome-wide copy number analyses revealed a trend toward increased tumor aneuploidy in tumors of non-responding patients (denoted by fraction of the genome with complete allelic imbalance; FDR-adjusted p = 0.19). AI, allelic imbalance; BOR, best overall response; CNV, copy number variation; Conseq, mutation consequence; CR, complete response; OS, overall survival; PD, disease progression; PR, partial response; SBS, single-base substitution; SD, stable disease. Dots represent hotspot mutations, and X denotes monoallelic loss of the wild-type allele.