Figure 6.
Multi-parameter Integrative Modeling Accurately Predicts Therapeutic Outcome
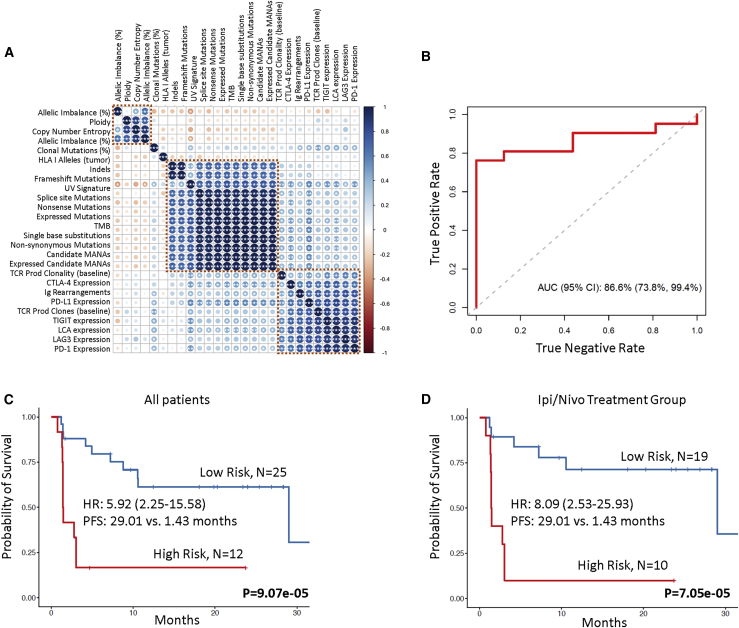
(A) Non-parametric correlations among genomic, transcriptomic, and T cell repertoire features were assessed by the Spearman’s rho statistic, and p values were corrected for multiple comparisons. The color of each dot refers to the Spearman rho coefficient value (darkest blue being 1 and darkest red being −1), and the size of each dot is proportional to the strength of the correlation. ∗∗∗FDR-adjusted p < 0.05, ∗∗FDR-adjusted p < 0.1, and ∗FDR-adjusted p < 0.2.
(B) Random forests were employed to integrate genomic, transcriptomic, and TCR features following training and testing with 10-fold cross-validation and resulted in an integrated model with an area under the curve of 0.866.
(C) Immunoglobulin rearrangements, TCR productive clones, PD-L1 expression, and expressed mutation load were combined in a multivariable Cox proportional hazards regression model, and a risk score was calculated for each case based on the weighted contribution of each parameter. The second tertile of the risk score was used to classify patients in high-risk (top 33.3%) and low-risk (bottom 66.6%) groups. Patients with a higher risk score had a significantly shorter progression-free survival compared to patients at low risk for disease progression (median PFS 1.43 months versus 29.01 months; log rank p = 9.07e−05; HR = 5.92; 95% CI: 2.25–15.58).
(D) This outcome was also pronounced in the ipilimumab/nivolumab group (median PFS 1.43 months versus 29.01 months for high- and low-risk score patients, respectively; log rank p = 7.05e−05; HR = 8.09; 95% CI: 2.53–25.93).