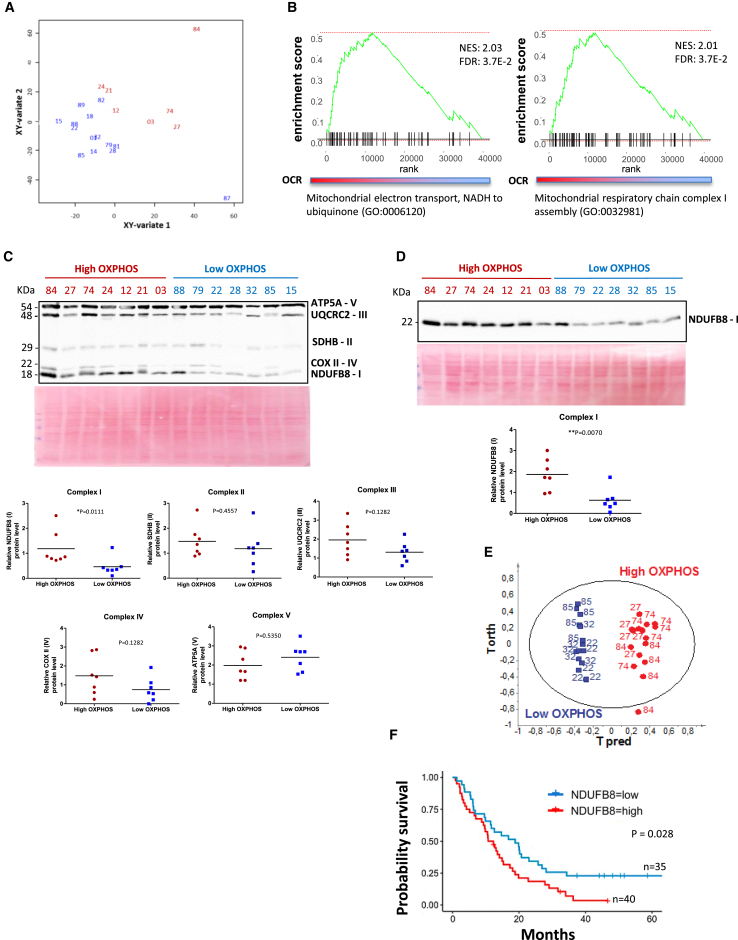
Figure 3.
Correlation between the High OXPHOS Status and the Abundance of Mitochondrial Complex I, Both at mRNA and Protein Level in the Primary PDAC Cancer Cells
High OXPHOS patients are depicted in red, and low OXPHOS are in blue.
(A) Canonical correlation analysis: graphical representation of PDX using the averaged components 1 and 2 of transcriptomic and metabolic datasets that were retrieved from canonical PLS.
(B) Gene set enrichment analysis: the two plots show the enrichment score for the related mitochondrial pathway on the second canonical PLS component in (A).
(C) Representative immunoblot showing the abundance of the 5 mitochondrial proteins (ATP5A, UQCR2, SDHB, COXII, and NDUFB8) belonging to mitochondrial complexes (V, III, II, IV, and I, respectively). Time exposure of the blot was 1 min. Ponceau red staining was used as the control of equal protein loading. Mitochondrial complexes protein levels were quantified from western blotting (WB) by ImageJ and normalized to Ponceau red staining. p value from Mann-Whitney test.
(D) Representative immunoblot showing the abundance of the mitochondrial protein NDUFB8 belonging to mitochondrial complex I. Time exposure of the blot was 1 min. Complex I protein was quantified by ImageJ and normalized to Ponceau red staining used as control of equal protein loading. p value from Mann-Whitney test.
(E) Metabolic analysis by HRMAS-NMR for 6 primary PDAC cancer cells derived from PDX, 3 high OXPHOS (27, 74, and 84) and 3 low OXPHOS (22, 32, and 85). OPLSDA score plot showing the discrimination between high OXPHOS and low OXPHOS.
(F) Kaplan-Meier survival curve using transcriptomic analysis on patient-derived xenografts, divided into high and low NDUFB8 gene expression groups (n = 40 and n = 35, respectively; the cutpoint was 13.22). p value was calculated by the log-rank test.
See also Figures S5 and S6 and Table S2.