Figure 3.
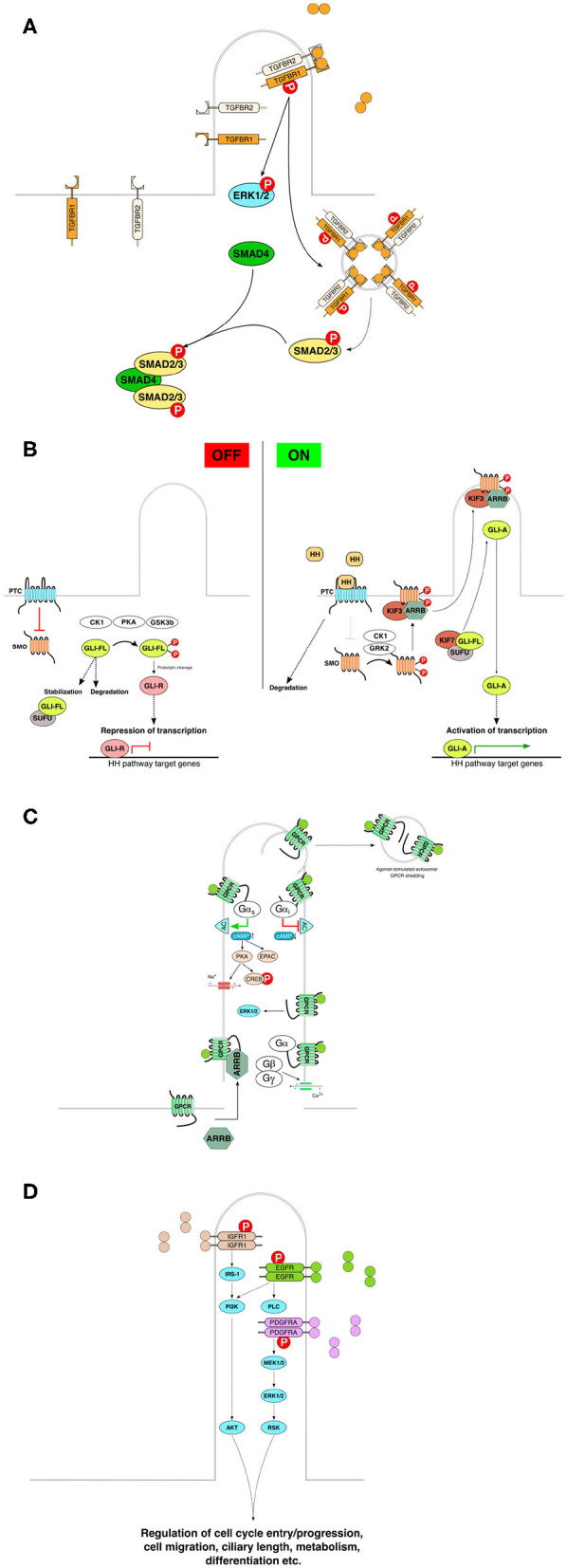
Basic components and rough characteristics of each indicated signaling pathway in relation to primary cilia. (A) TGFbeta receptors accumulate in the primary cilia, and ligand binding to the TGFBR2 faciliates recruitment of TGFBR1 to form the heterodimeric receptor complex. Subsequent phosphorylation and activation of the receptor leads to downstream activation of the SMAD family of transcription factors by active receptor transport and aggregation in endosomal vesicles for downstream signal enhancement. (B) Mammalian Hedgehog signaling is tightly linked to the primary cilia. In the absence of signal, the pathway is maintained inactive or in a repressive state through the inhibitory action of Patched on Smoothened, and through the proteolytic cleavage or SUFU-mediated stabilization of GLI family of transcription factors. Upon activation by Hedgehog pathway ligands (Sonic Hedgehog, Indian Hedgehog or Desert Hedgehog) the inhibitory activity of PTC on SMO is abrogated and SMO can, with the help of beta-arresin and kinesin 3, translocate to the primary cilia. Active SMO then facilitates dissociation of GLI from its regulator SUFU leading to activation of GLI and GLI-A-induced transcription of HH pathway response genes in the nucleus. (C) The primary cilia-associated G-protein-coupled receptors are transported to the primary cilia assisted by e.g., beta-arrestin. Ligand stimulation leads to receptor activation and activation of downstream signaling depending on the receptor's coupling to various types of G-proteins. Ligand-induced shedding of G- proteins in ectosomes from the primary cilia denotes an additional level of regulation or suppression of G-protein signaling. (D) Receptor tyrosine kinase pathways are also associated with the primary cilia. For example, IGF, EGF, and PDGF utilize primary cilia for signaling. Ligand binding results in downstream activation of specific kinase cascade activations leading to activation of transcription factors and intracellular effectors that participate in the regulation of various cellular processes. Redrawn using information from the following: Ruel and Thérond (2009), Ishikawa and Marshall (2011), Christensen et al. (2012), Hilgendorf et al. (2016), Bangs and Anderson (2017), Christensen et al. (2017), and Anvarian et al. (2019). BBsome, Bardet-Biedl syndrome protein complex; IFT, intraflagellar transport; PTC, patched; SMO, smoothened; SUFU, suppressor of fused homolog; GLI-FL, glioma-associated oncogene (zinc finger protein GLI) full-length; GLI-A, activated form of GLI; GLI-R, repressor form of GLI; CK1, casein kinase 1; PKA, protein kinase A; GSK3b, glycogen synthase kinase 3 beta; HH, hedgehog; GPRK2, G-protein- coupled receptor kinase 2; ARRB, beta-arrestin; KIF, kinesin superfamily protein; TGFB, transforming growth factor beta; TGFBR, TGFB receptor; SMAD, family of transcription factors homologous to C. elegans SMA and MAD families; ERK, extracellular signal-regulated kinase; MEK, mitogen-activated protein kinase kinase; IGFR, insulin-like growth factor receptor; EGFR, epidermal growth factor receptor; PDGFRA, platelet-derived growth factor receptor alpha; IRS, insulin receptor substrate; PI3K, phosphoinositide 3-kinase; PLC, phospholipase C; AKT, protein kinase B; RSK, ribosomal protein S6 kinase; GPCR, G-protein-coupled receptor; AC, adenylate cyclase; cAMP, cyclic adenosine monophosphate; CREB, cAMP-responsive element-binding protein; EPAC, rap guanine nucleotide exchange factor, exchange protein directly activated by cAMP; G alpha s, G-protein alpha subunit, stimulatory; G alpha i, G-protein alpha subunit, inhibitory; Gbeta, gamma, G-protein beta and gamma subunits, respectively; RTK, receptor tyrosine kinase.
