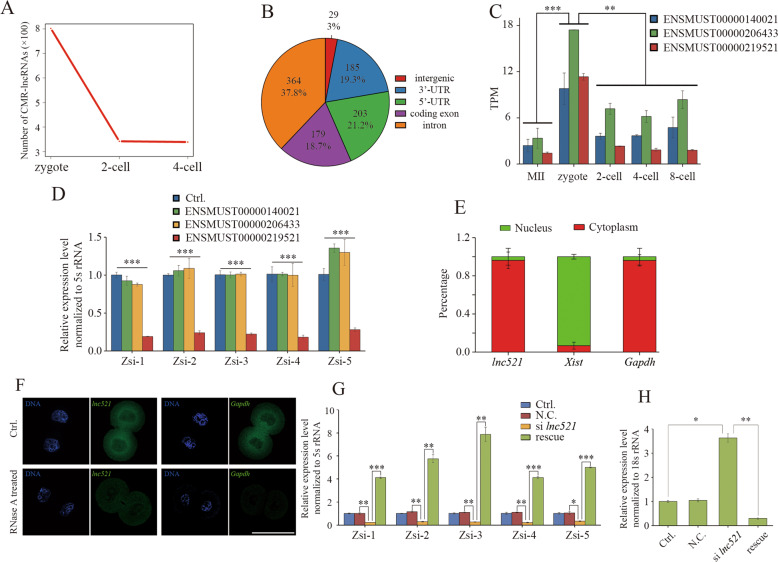
Fig. 5. Prediction and verification of CMR-lncRNAs in AGO2-dependent MRD.
A Number of CMR-lncRNAs predicted in zygotes, 2-cell embryos and 4-cell embryos. The number of lncRNA in zygote is significantly higher than that of 2-cell and 4-cell embryos. B Annotation of CMR-lncRNAs in early mouse embryos. CMR-lncRNAs were annotated using all available annotation sources, and a total of 37.8 and 18.7% were found residing in introns and coding exons, respectively. C Expression patterns of ENSMUST00000140021, ENSMUST00000206433, and ENSMUST00000219521 according to RNA-seq data in mouse early embryos. D Effect of ENSMUST00000140021, ENSMUST00000206433 and ENSMUST00000219521 knockdown on the expression of Zfp277 endosiRNAs. The results show that deletion of ENSMUST00000219521 (lnc521) impaired the biogenesis of endosiRNAs. Error bars indicate the s.e.m. E Subcellular localization of lnc521 by RNA fractionation and q-PCR analysis. Error bars indicate the s.e.m. F Representative images of RNA FISH show subcellular localization of lnc521 in 2-cell embryos (lnc521: n = 17; Gapdh: n = 13). RNase A-treated 2-cell embryos were tested as the N.C. (lnc521: n = 11; Gapdh: n = 13). Scale bar, 50 µm. G Expression levels of Zfp277 endosiRNAs measured by q-PCR in 2-cell embryos of the control, si lnc521 injection and si lnc521 injection with lnc521 RNA add back. Error bars indicate s.e.m. H Expression levels of Zfp277 measured by q-PCR in 4-cell embryos of control, si lnc521 injection and si lnc521 injection with lnc521 RNA add back (lnc521 rescue). *p < 0.05; **p < 0.01; ***p < 0.001. Error bars indicate the s.e.m.