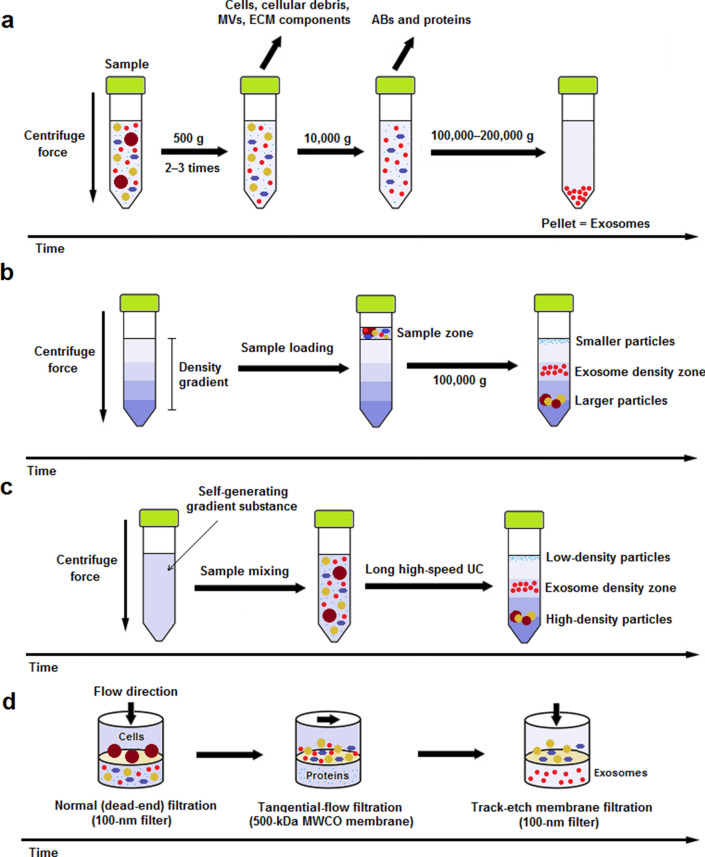
Fig. 1.
Schematic representation of most frequently utilized exosome isolation methods for therapeutic purpose. a Differential ultracentrifugation (DUC): Sample is subjected to 2‒3 steps of low-speed (500 g) centrifugation to pellet out cells, microvesicles (MVs), extracellular matrix (ECM) components, and cellular debris. The supernatant is then centrifuged at 10,000 g for removal of apoptotic bodies (ABs) and contaminating proteins. Finally, exosomes are retrievd by a long (60–120 min) ultracentrifugation (UC) step at 100,000–200,000 g and subsequent washing of the pellet in PBS; b rate-zonal ultracentrifugation (RZUC): RZUC is a type of density gradient UC (DGUC) where sample is placed at the surface of a gradient density medium such as sucrose, and following a step of UC at 100,000 g, sample components migrate through the gradient density and separate according to their size and shape; c isopycnic ultracentrifugation (IPUC): IPUC is another type of DGUC that separates particles based on their density. Sample is usually mixed with a self-generating gradient substance such as CsCl, and is then subjected to a long UC step. In the end, distributed components form bands, so-called the isopycnic position, where the buoyant density of the collected particles matches with the gradient density of the surrounding solution. The banded exosomes can be retrieved from the density zone between 1.10 and 1.21 g/mL by fractionation; d sequential filtration (SF): Sample is first subjected to a 100-nm dead-end (normal) filteration process to separate cells and larger particles. Then, contaminating proteins are excluded via tangential flow filtration using a 500-kDa MWCO membrane. Lastly, the filtrate is once more passed through a track-etch membrane filter (with pore size of 100 nm) at very low pressure in order to inhibit passing of flexible nonexosomal EVs into the filtrate while allowing for passage of exosomes