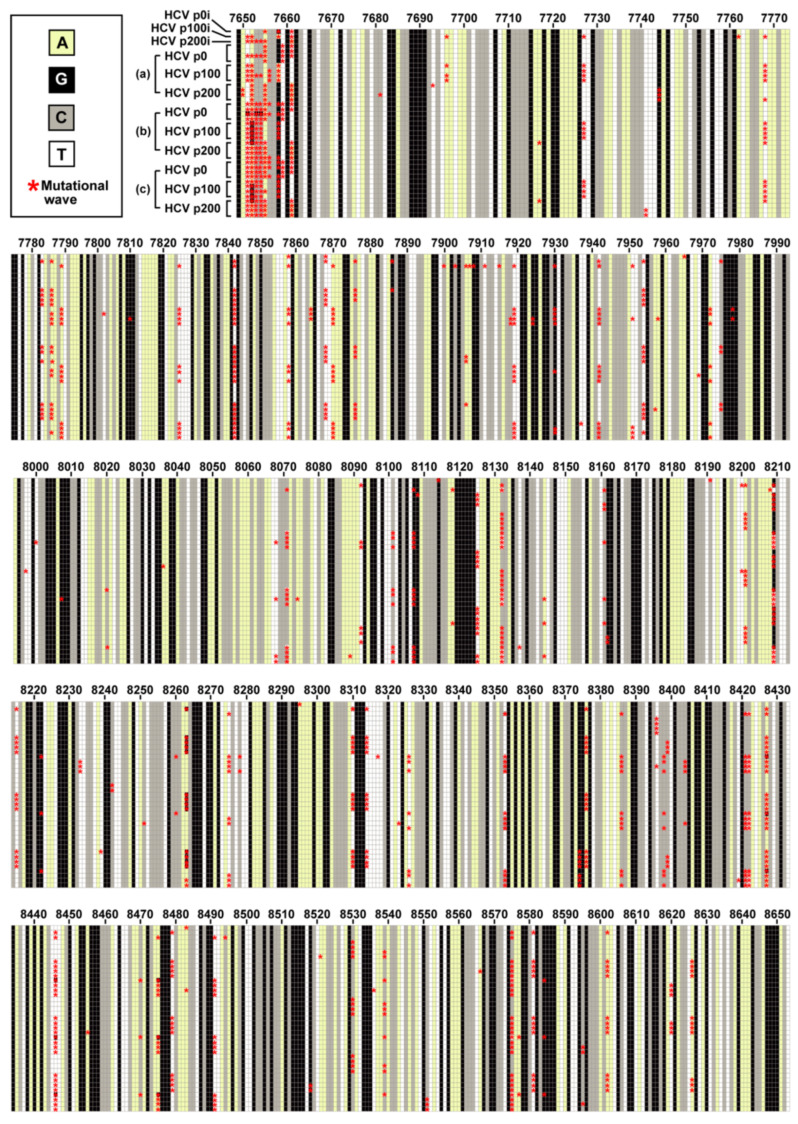
Figure 2.
Heat map of consensus sequences of 39 HCV populations derived from HCV p0 upon passage in Huh-7.5 cells. The region analyzed by deep sequencing spans genomic residues 7649 to 8653 (residue numbering according to reference isolate JFH-1). The populations are those identified by empty circles in the HCV passage diagram of Figure 1A, and are indicated at the left of the top bloc of the alignment; (a), (b), and (c) refer to the three replicas of the four serial passages of HCV p0, HCV p100 and HCV p200. Each horizontal alignment of squares displays the consensus sequence (1005 nucleotide positions) of the population written on the left, with the nucleotide color code given in the upper box on the left. The red asterisks indicate the nucleotides that participated in mutational waves (that is, that changed in frequency among any of the populations analyzed). The complete set of mutations and deduced amino acid substitutions are given in Table S1.