Figure 4.
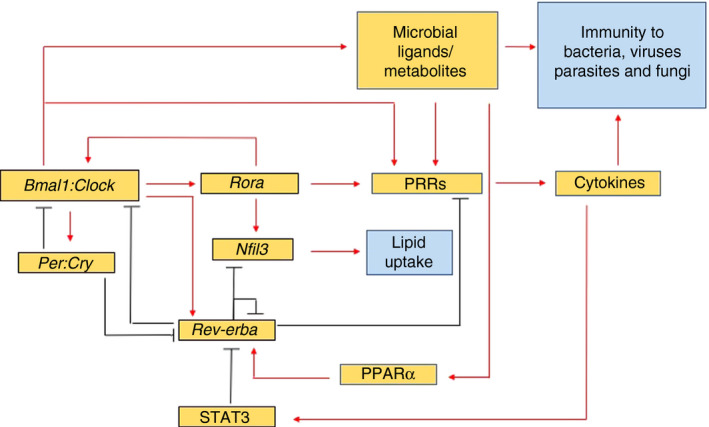
Microbial modulation of circadian rhythms. PRR oscillation (referring to published work in TLRs and Nlrp3) is driven by competition between the Rev‐erbα repressor and the Rorα activator, both of which directly bind and compete for the ROR response element, a DNA binding sequence, present in Bmal1. This competition promotes the oscillation of antimicrobial responses, for example cytokines, which are mediated by downstream NFκB and AP‐1 binding to target genes. Bmal1 binds to an Ebox element (a DNA sequence) in Rev‐erbα activating its transcription, while a DR2 element (another DNA sequence) in Rev‐erbα mediates its autorepression and activation by PPARα. DNA elements (RORE, Ebox, Dbox – not shown) are present in TLR genes, Nod2, circadian genes and many other genes, including many which influence immune processes. Circadian regulation of Nfil3 by the Rev‐erbα repressor and the Rorα activator can also modulate intestinal lipid absorption and transport 23 . Another mechanism of circadian regulation of TLRs has also been identified in peritoneal macrophages, whereby Tlr9 expression is controlled by direct binding of Bmal1 to the Ebox sequence present in the Tlr9 gene 80 . Red lines indicate activation; black lines indicate inhibition. Blue boxes indicate the known effects of microbiota in modulating circadian rhythms.
