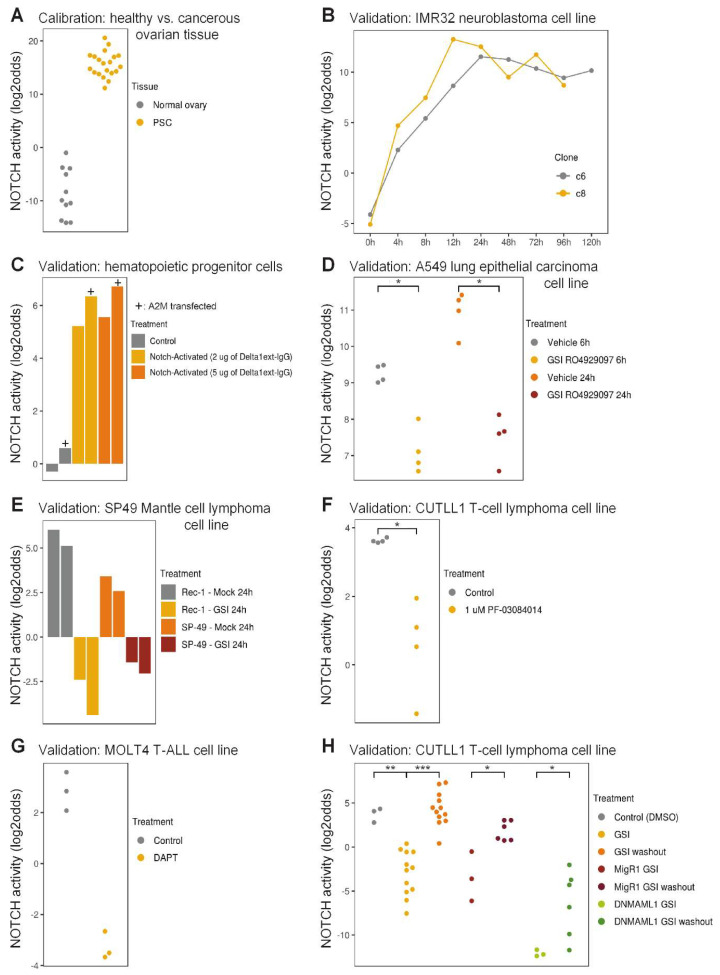
Figure 1.
Calibration and biological validation of the Notch pathway model. (A) Calibration of the Notch pathway model. GSE7307, GSE18520 [90], GSE29450 [91], GSE36668 [92], normal ovarian tissue samples (inactive pathway); GSE2109, GSE9891 [93], high grade serous ovarian cancer samples (active pathway). (B–G) Validation of the model on independent GEO datasets from different cell lines. * p < 0.05, ** p < 0.01, *** p < 0.001. (B) GSE16477 [94]. Two clones (c6 and c8) of the IMR32 neuroblastoma cell line at different times (0–120 h) after induction of active intracellular NOTCH3. (C) GSE29524. A2M (+ symbol) or control vector-transfected CD34+CD45RA-Lin- hematopoietic progenitor cells from umbilical cord blood were cultured for 72 h on a surface with 0, 2 or 5µg plastic-immobilized NOTCH ligand Delta1ext-IgG. A2M is a nuclear-trapped mutant of AF1q/MLLT11. (D) GSE36176 [95]. A549 lung cancer cell lines subjected to vehicle control or gamma secretase inhibitor (GSI) RO4929097 for 6 or 24 h. (E) GSE34602 [89]. Rec-1 (containing an activating NOTCH1 mutation) and SP49 Mantle cell lymphoma cell lines subjected to vehicle control or GSI compound E for 24 h. SP49 cells harbor an activating NOTCH4 rearrangement. (F) GSE33562 [96]. Duplicate samples of the CUTLL1 T-cell lymphoma cell line were subjected to vehicle control or the GSI PF-03084014 (1µM) for 48 h. (G) GSE6495 [97]. MOLT4 T-cell acute lymphoblastic leukemia cell line before and 48 h after addition of the GSI DAPT (5µM); three independent experiments. (H) GSE29544 [82]. CUTLL1 T-cell lymphoblastic lymphoma cells subjected to GSI compound E (1 µM) for 3 days. From left to right: DMSO control; 3 grouped conditions: GSI without or with 2 or 4 h mock washout; 4 grouped conditions: GSI followed by 2 or 4 h GSI washout in the presence or absence of cycloheximide; GSI in presence of a control viral transcript MigR1; 2 grouped conditions: GSI in the presence of a control viral transcript MigR1 with 2 or 4 h washout; GSI in the presence of viral transcript DNMAML1; 2 grouped conditions: GSI in the presence of viral transcript DNMAML1 with 2 or 4 h washout. The activity score is calculated as log2odds. Two-sided Wilcoxon signed-rank statistical tests were performed, p-values are indicated in the figures. In case fewer than 3 samples were needed for presentation, bar plots are used instead of dot plots.