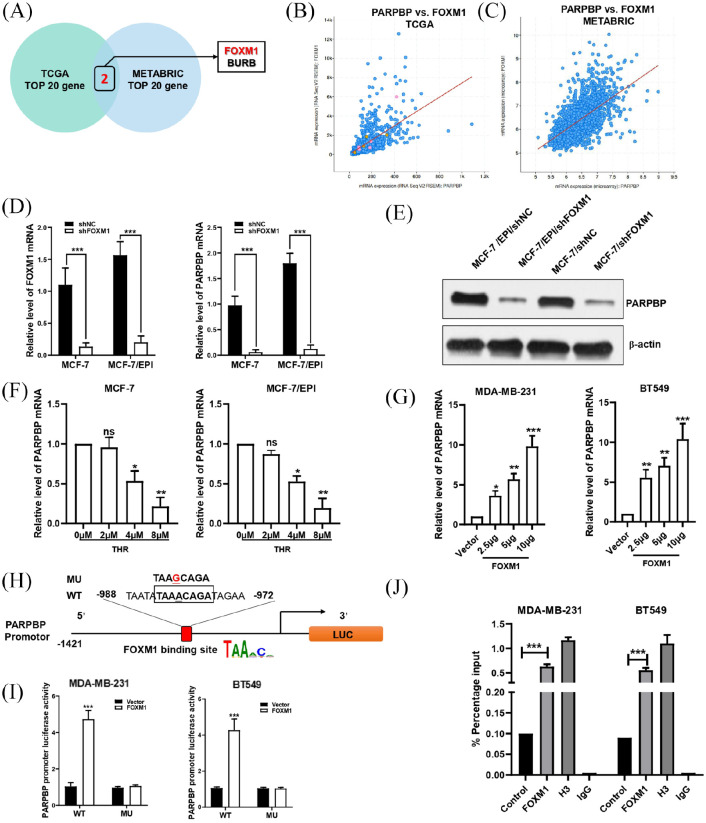
Figure 6.
FOXM1 directly binds to the PARPBP promoter and regulates its activity. (A) Venn diagram shows that FOXM1 and BURB are the intersection of the top 20 PARPBP co-expressed genes in the TCGA and METABRIC datasets. (B, C) Correlation between FOXM1 and PARPBP in TCGA (B) and METABRIC (C). (D, E) Downregulation of FOXM1 by transfecting shFOXM1 reduced PARPBP expression by qRT-PCR (D) and western blotting (E). (F) MCF-7 and MCF-7/EPI cells treated with FoxM1 inhibitor THR at concentrations of 0, 2, 4, and 8 μM. (G) Overexpression of FOXM1 by transfecting FOXM1 expression plasmid in MDA-MB-231 and BT549 cells increased the mRNA level of PARPBP. (H) Schematic of the PARPBP promoter reporter and its putative FOXM1-binding site. (I) Luciferase reporter assay in MDA-MB-231 and BT549 cells (with FOXM1 expression plasmid or empty vector) transfected with luciferase reporter constructs containing WT or MU PARPBP promoter. Data represent means ± SD of at least three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (J) ChIP assay showing the binding of FOXM1 to PARPBP promotor. Chromatins were isolated from MDA-MB-231 and BT549 cells, and specific primers for PARPBP promotor was used to DNA quantification. The enrichment percentage = 2% × 2[CT (input sample) − CT (IP sample)]. Normal IgG and histone H3 were used as negative and positive controls, respectively.
ChIP, chromatin immunoprecipitation; GEPIA, Gene Expression Profiling Interactive Analysis; IgG, immunoglobulin G; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; MU, mutant; OS, overall survival; PARBP, PARP1 binding protein; SD, standard deviation; THR, thiostrepton; TCGA, The Cancer Genome Atlas; TPM, transcripts per million; WT, wild type.