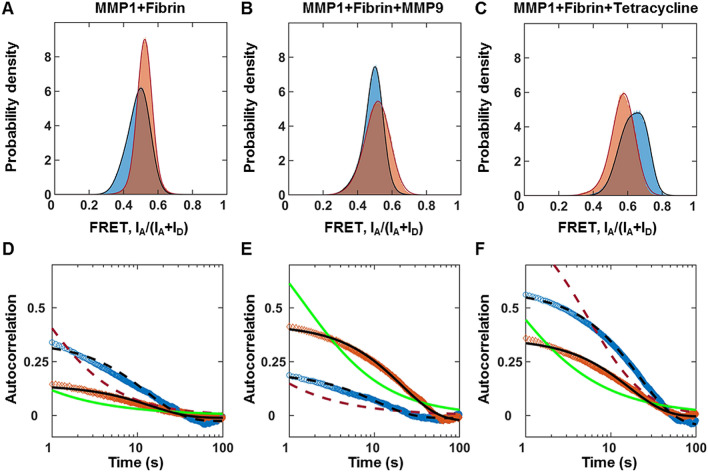
Figure 3.
Interdomain dynamics of MMP1 on fibrin correlates with activity. More than 300,000 smFRET values at 22 °C with a 100 ms time resolution for each condition are used to create area-normalized histograms of MMP1 interdomain distance (bin size = 0.005). (A) Histograms without ligand, (B) Histograms in the presence of MMP9 (an enhancer), and (C) Histograms in the presence of tetracycline (an inhibitor) for active (blue) and active site mutant (orange) MMP1. Histograms are fitted to a sum of two Gaussians (active: solid blue line; active site mutant: solid red line). Autocorrelations of MMP1 interdomain distance are calculated from the time series of smFRET values. (D) Autocorrelations without ligand, (E) Autocorrelations in the presence of MMP9, and (F) Autocorrelations in the presence of tetracycline for active MMP1 (blue) and active site mutant of MMP1 (orange). Autocorrelations are fitted to exponentials and power laws (exponential fit to active: dashed black line; power law fit to active: dashed red line; exponential fit to active site mutant: solid black line; power law fit to active site mutant: solid green line). The error bars in the histograms and autocorrelations represent the square roots of the bin counts and the standard errors of the mean (sem) and are too small to be seen. The supplementary information contains the fit equations and the best-fit parameters for histograms and autocorrelations (Table S1). We approximated smFRET efficiency by IA/( IA + ID)49 and has no unit.