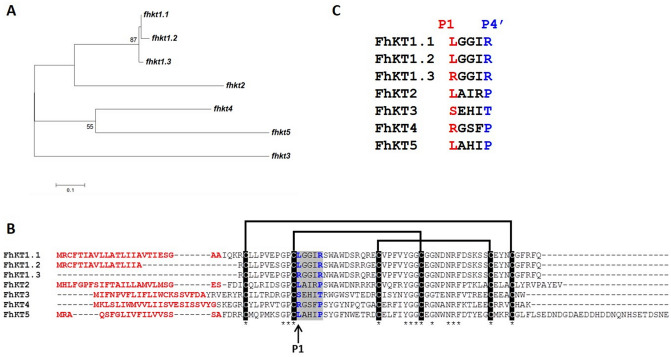
Figure 1.
A family of Kunitz-type inhibitors is present in the F. hepatica genome. (A) Maximum-likelihood phylogenetic tree based on sequences encoding the kunitz domain in F. hepatica genome. Bootstrap values > 50% from 1000 replicate iterations are shown. (B) Amino acid sequence alignment of the translated amino acid sequence of each fhkt gene. The P1, residue 15, within the reactive site loop is indicated with an arrow. Secretory signal peptides are shown in red and lines show the three conserved disulphide bonds that occur between the six conserved cysteine residues, highlighted in black, specifically between Cys1 and Cys6, Cys2 and Cys4, and Cys3 and Cys5. The Kunitz reactive loop region is shaded in grey and P1 and P4′ residues predicted to interact with residues at the S2 and S2′ sites of cathepsin L-like cysteine proteases are highlighted in blue. (C) Amino acid sequence alignment of the reactive loop region of the seven members of the Kunitz family. The P1 and P4′ residues are highlighted red and blue, respectively.