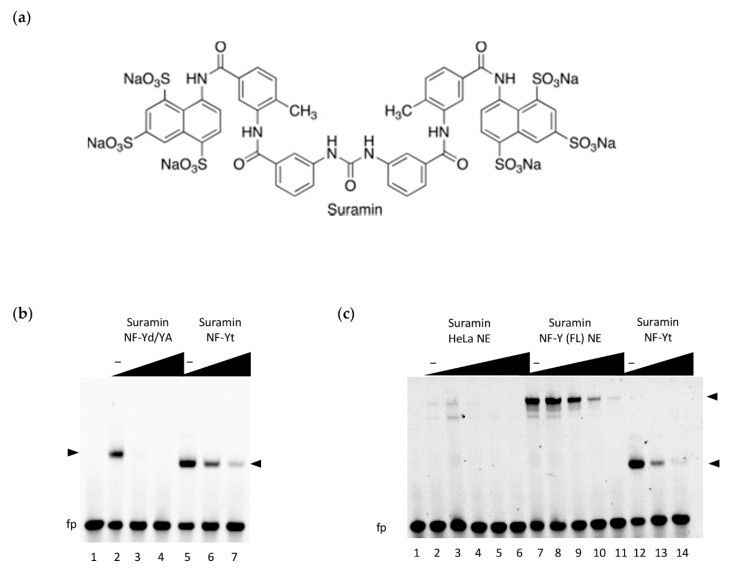
Figure 1.
DNA-binding inhibition by suramin; (a) chemical structure of suramin; (b) DNA binding inhibition of the NF-Y trimer (40 nM) by suramin was assessed by electrophoretic mobility shift assays (EMSA) using a Hsp70 CCAAT box DNA probe (20 nM). Inhibition was tested at increasing doses of suramin (0, 50, 100 µM; lanes 2–4 and 5–7) against the reconstituted trimer, obtained by mixing equimolar ratios of purified NF-YA with NF-Y histone fold domain (HFD) dimer (NF-Yd) (NF-Yd/YA) or on the co-purified trimeric subunit protein (NF-Yt). Lane 1: probe alone DNA binding mix in the absence of NF-Y. NF-Y/DNA complexes are indicated by arrowheads. fp: free probe. Slower migration of NF-Yd/YA/DNA, as compared to NF-Yt/DNA (composed of the minimal DNA-binding domains), reflects the higher molecular weight of the purified NF-YA subunit within the complex; (c) EMSA experiments using nuclear extracts (NE) from HeLa cells, obtained from control cells (HeLa NE: lanes 2–6) or from cells overexpressing the full-length NF-Y subunits (including the transactivation domains) (NF-Y(FL) NE: lanes 7–11). NF-Yt recombinant protein was used as a positive control for suramin inhibition of DNA binding (lanes 12–14). In lanes 2–11, the reaction mix includes 2.3 µg of nuclear extract, and an increasing concentration of suramin for NE and NE + NF-Y(FL) (0, 1, 50, 100, and 200 µM), lanes 12-14 reactions include NF-Yt protein (40 nM) and suramin (0, 100, and 200 µM). Lane 1: probe alone DNA binding mix without NF-Y subunits or NE added. The migration of FL and minimal domain NF-Y/DNA complexes are indicated by black arrowheads. “fp”: free probe, “‒”: no suramin.