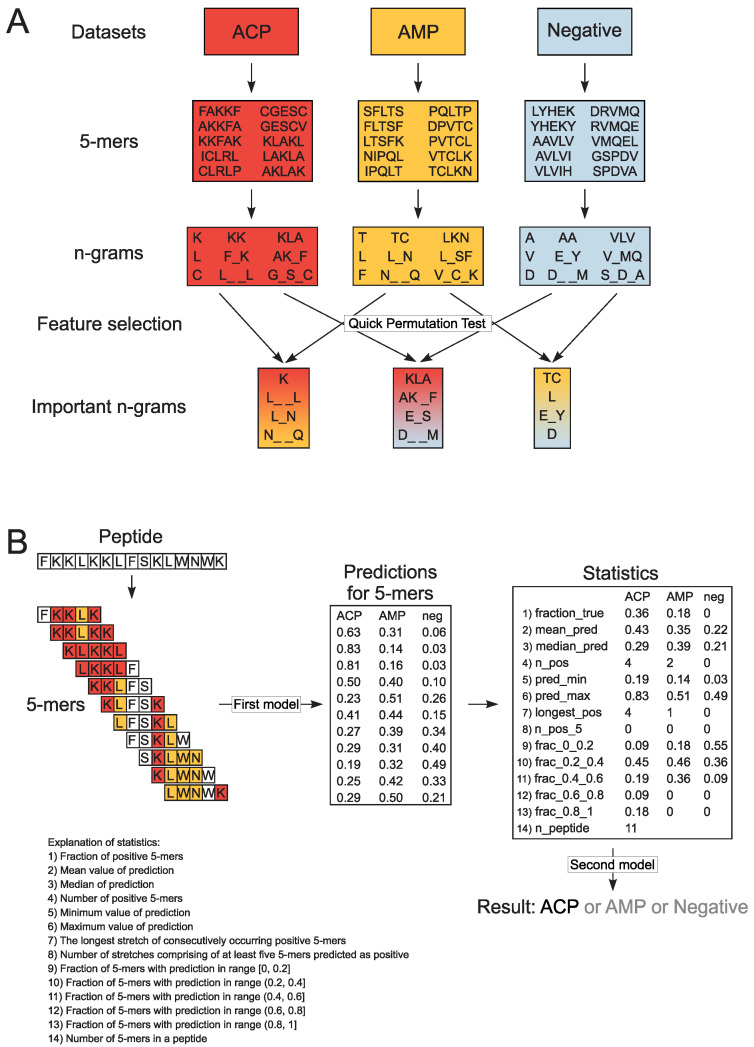
Figure 1.
Schematic representation of n-gram extraction (A) and decision-making procedure in CancerGram (B). The training data sets include ACP (shaded in red), AMP (shaded in yellow) and non-ACP/non-AMP sequences (the negative data set, shaded in blue). Each peptide from the training data sets was divided into subsequences of 5 amino acids (5-mers). For each 5-mer, we extracted continuous and discontinuous n-grams of size ranging from 1 to 3, and exemplary n-grams are presented in boxes shaded in colors respective to the data sets. The informative n-grams for CancerGram training were selected by Quick Permutation Test for all combinations of the data sets, and they are shaded in: (i) red-yellow for the ACP/AMP data set, (ii) red-blue for the ACP/Negative data set, and (iii) yellow-blue for the AMP/Negative data set (A). To make a prediction, CancerGram first divides a peptide into 5-mers and then, for each 5-mer, makes a prediction if it is an ACP, AMP or non-ACP/non-AMP (the first model). To scale the prediction from 5-mers to the level of a peptide, numerous statistics are calculated, and on their basis, CancerGram makes the final prediction (the second model) (B).