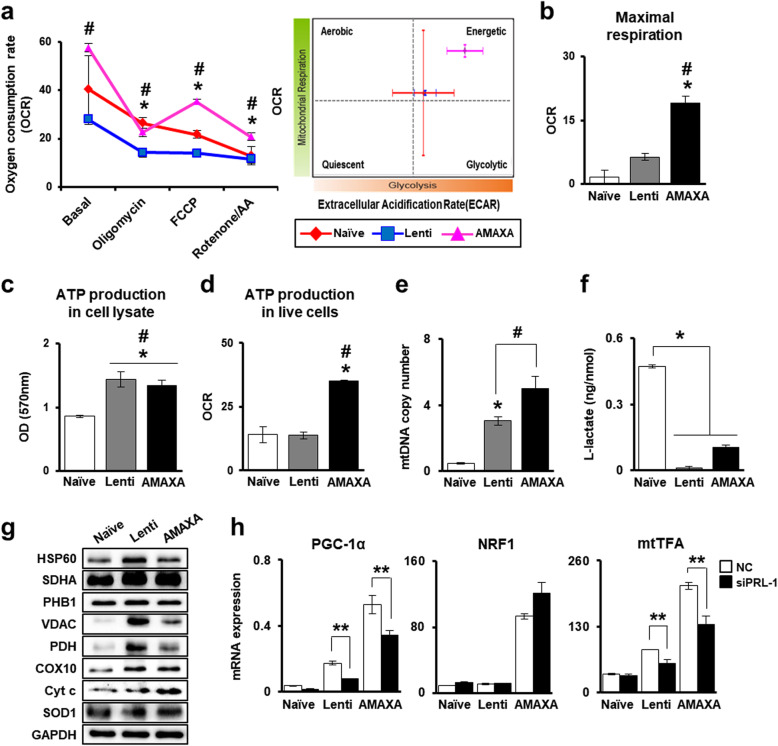
Fig. 4.
Improved mitochondrial respirational metabolic states of PD-MSCsPRL-1 regulate mitochondrial dynamics. a OCR and ECAR of PD-MSCsPRL-1 using lentiviral and nonviral AMAXA systems after sequential treatment with 1 μM oligomycin, 0.5 μM FCCP, and 0.5 μM rotenone/AA by the Seahorse XF24 Analyzer. Quantification of b maximal respiration and d ATP production under live conditions of naïve and PD-MSCsPRL-1. c ATP production assay in naïve and PD-MSCPRL-1 lysate (10 μg/μl). e mtDNA copy number of PD-MSCsPRL-1 using lentiviral and nonviral AMAXA systems by the TaqMan assay. Human nuclear DNA was used as an internal control. f L-lactate production assay of PD-MSCsPRL-1 using cell lysate (< 25 mg/ml). g Western blotting of mitochondrial metabolism-specific genes in naïve and PD-MSCsPRL-1. h qRT-PCR analysis of mitochondrial biogenesis-specific genes (PGC-1α, NRF1, and mtTFA) in PD-MSCPRL-1-treated or -untreated siRNA-PRL-1 (siPRL-1; 50 nM) after 24 h. Data from each group are shown as the mean ± SD. *p < 0.05 versus naïve; #p < 0.05 versus lenti; **p < 0.05 versus NC. AA, antimycin A; COX10, cytochrome c oxidase 10; Cyt c, cytochrome c; ECAR, extracellular acidification rate; FCCP, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; HSP60, heat shock protein 60; mtTFA, mitochondrial transcription factor A; NC, negative control; NRF1, nuclear respiratory factor 1; OCR, oxygen consumption rate; PDH, pyruvate dehydrogenase; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator-1 alpha; PHB1, prohibitin 1; SDHA, succinate dehydrogenase; SOD1, superoxide dismutase 1; VDAC, voltage-dependent anion channel