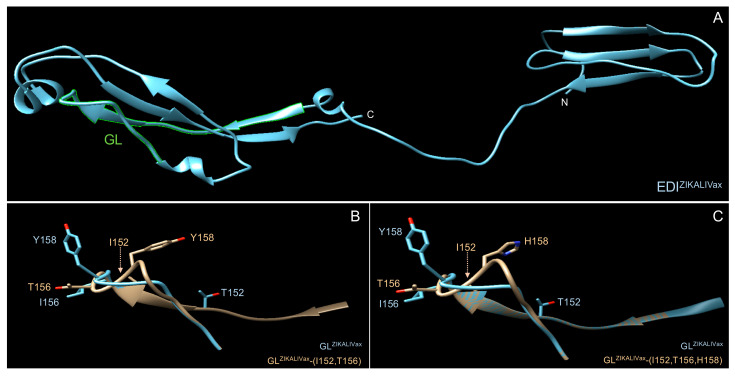
Figure 7.
Structural features of rEDIZIKALIVax. Tridimensional structure prediction of rEDI ZIKALIVax and its mutants was performed by comparative modeling on PHYRE2 protein recognition server (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index). The predicted structures were analyzed with Chimera, a program for interactive visualization of tridimensional molecules. In (A), predicted structure of ZIKALIVax rEDI with GL region in green. The two C-terminal tags of rEDI are not presented. In (B,C), the ZIKALIVax GL region structure (GLZIKALIVax) was compared to GL mutants bearing the two mutations E-T152I, and E-I156T (B) or the three mutations E-T152I, E-I156T, and E-Y158H (C). The hatched arrow indicates the position of residue I152 in the 3D structure of GL mutant.