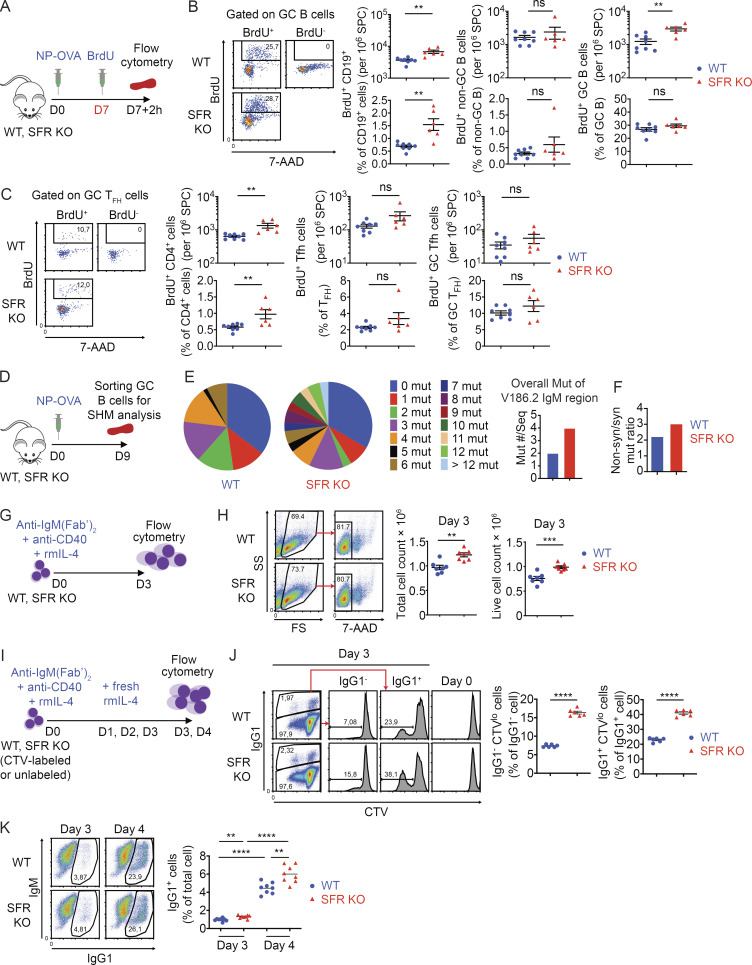
Figure 5.
SFR KO GC B cells display increased proliferation, SHM, and isotype switching. (A–C) As in Fig. 2 B, except that proliferation was assessed in vivo using BrdU incorporation. The protocol is outlined in A, whereas BrdU incorporation in total B cells (CD19+), non-GC B cells, GC B cells, total CD4+ cells, Tfh cells, and GC Tfh cells is depicted in B and C (WT, n = 8; SFR KO, n = 6). Data are pooled from three independent experiments. Each dot represents one mouse. (D–F) After immunization with NP-OVA, GC B cells were purified from three independent mice of each genotype. mRNA was isolated, cDNAs were synthesized, and the V186.2 region of the IgM-encoding gene was sequenced. The protocol is shown in D, and analyses of the sequencing data are shown in E and F. The proportions of cDNA sequences with the indicated numbers of mutations are depicted by pie charts (E, left). The overall average numbers of mutations per sequence are shown (E, right). The ratios of nonsynonymous over synonymous mutations are shown (F). The numbers of sequences analyzed were WT, n = 65 and SFR KO, n = 70. Data were pooled from two experiments. For statistical analysis of mutation numbers, a χ2 statistical analysis was performed on pooled data comparing the distribution of mutated sequences (i.e., the proportions of mutations with 0, 1, 2, etc. mutations per sequence) between WT and SFR KO mice. As χ2 requires that all expected values be >1.0 and at ≥20% of the expected values be >5, we pooled the more highly mutated sequences (5–10 mutations per sequence) into a bin for each genotype. The calculated P value was 0.0006 and is shown above the pie chart. (G and H) Purified naive B cells were seeded in activation medium containing anti-IgM(Fab’)2, anti-CD40 and recombinant mouse (rm) IL-4 (G). On day 3, total and live cells were enumerated (H). Data are pooled from four independent experiments. Each dot represents one mouse. (I–K) Purified naive B cells from WT (n = 2) or SFR KO (n = 2) mice were labeled or not labeled with CTV and activated in vitro as in G. The protocol is depicted (I). At day 3, CTV-labeled cells were analyzed for dividing cells (CTVlo) in IgG1− and IgG1+ subsets (J). Assays were in triplicate. At day 3 or 4, unlabeled cells were assayed for IgG1+ cells (K). Assays were in quadruplicate. Data are representative of two experiments. Each symbol represents a different well from two independent mice. Data are representative of two experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.001 (two-tailed Student’s t test). Data are presented as mean ± SEM. D, day; mut, mutations; ns, not significant; seq, cDNA sequences; syn, nonsynonymous.