FIGURE 3.
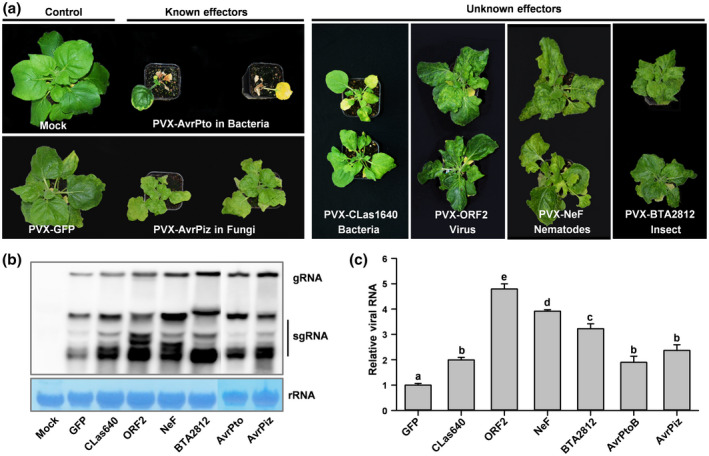
Confirmation and identification of virulence effectors in multiple plant pathogens using the virus‐induced virulence effector (VIVE) assay. (a) Analysis of two known and four novel virulence effectors from six plant pathogens in leaves of 10–14‐day‐old Nicotiana benthamiana plants (n = 10) using the PVX‐based expression system. Leaves were infiltrated with Agrobacterium (OD600 = 0.6–0.8) carrying the PVX‐Effector construct, and photographs were taken at 21 days postinfiltration (dpi). (b) RNA blot analysis of the accumulation of PVX genomic and subgenomic RNAs in N. benthamiana plants at 21 dpi. Total RNA was extracted from infected N. benthamiana leaves at 21 dpi. Wild‐type N. benthamiana plants (mock) were used as a negative control. (c) Relative viral RNA accumulation in plants inoculated with the PVX‐GFP or PVX‐Avh148 construct. Analysis of the PVX CP gene by quantitative reverse transcription PCR (RT‐qPCR) was used to determine the viral RNA level. Data represent mean ± SE of three independent experiments. The EF‐1α gene was used as an internal control. Each experiment was performed twice, with similar results. Different letters indicate statistically significant differences among samples (p < .01; Duncan's multiple range test)
