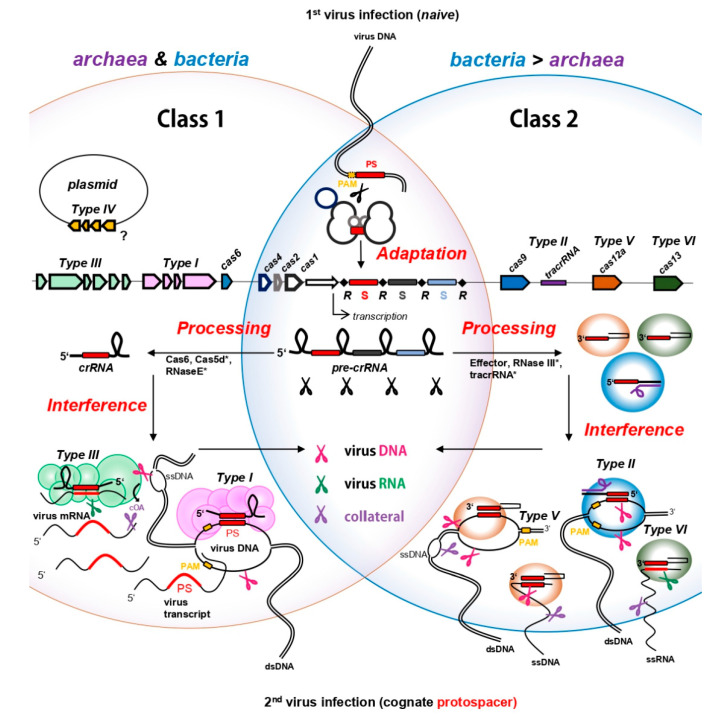
Figure 1.
CRISPR types in prokaryotes and their mechanisms of action. Class 1 systems (including types I, III, and IV) are prevalent in both archaea and bacteria, whereas Class 2 systems (including types II, V and VI) are found almost exclusively in bacteria. Mechanisms and genes in the overlapping region are shared by the two classes. The different CRISPR steps are indicated in red and explained in the main text. Both classes contain effector complexes that can perform all three types of interference: virus DNA interference (indicated by pink scissors), virus RNA interference (indicated by green scissors) and collateral damage (i.e., promiscuous cleavage of host and virus DNA/RNA, indicated by purple scissors). “S” refers to “spacer”, “R” refers to “repeat”. * Asterisks refer to enzymes that are not involved in processing of all classes: Cas5d replaces Cas6 function in type I-C [25]; RNaseE is involved in processing in a type III-B system [26]; RNase III processes pre-crRNA in type II; tracrRNA is needed in type II and certain subtypes of type V (see text). Type I-D targets ssDNA and dsDNA (not shown) [27]; In type II systems, Csn2 is also involved in spacer acquisition (not shown); spacer acquisition from RNA via RTs is not shown [28]; ssRNA targeting of the type V-G subtype is not shown [29] (see text).