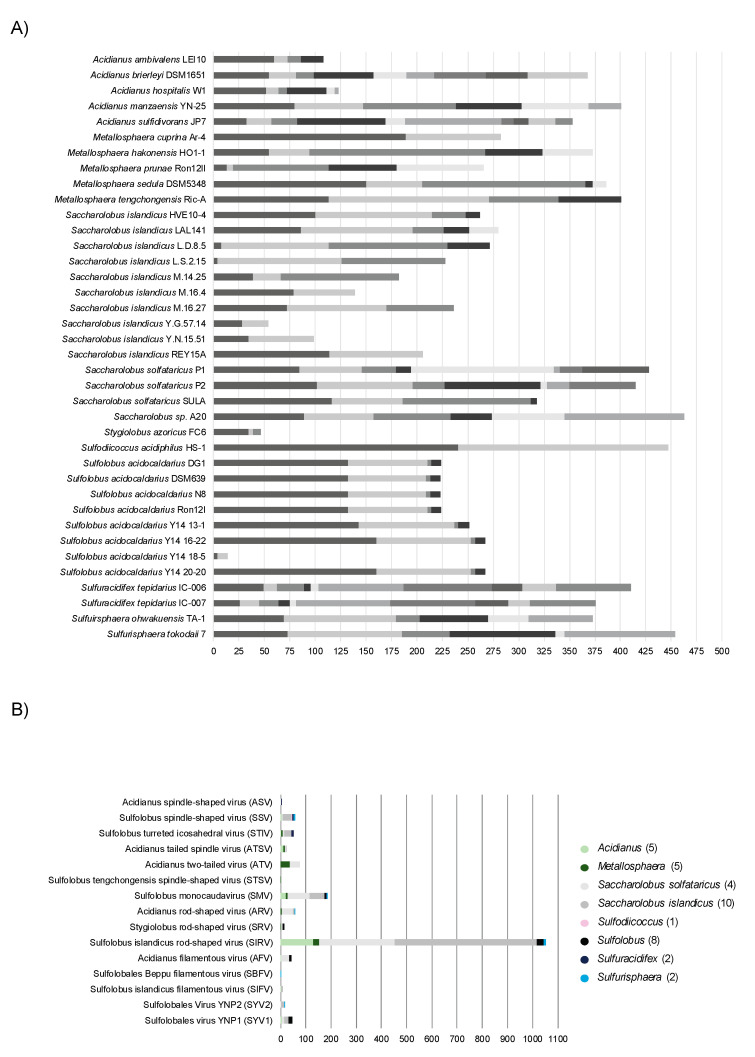
Figure 5.
CRISPR array, spacers and virus matches in Sulfolobales genera. (A) The number of arrays (depicted in different color shadings) and the number of spacers per array are shown for representative genomes of the Sulfolobales. (B) The stacked bar plot shows the total amount of spacers identified on the genus level that (partially) match protospacers carried on genomes of viruses associated with the Sulfolobales. The data (CRISPR arrays and spacer) for the analyses were retrieved using programs CRISPRCasFinder (CRISPR-Cas++ 1.1.2, [241]) and orientation of CRISPR arrays was determined using CRISPRstrand (implemented in CRISPRmap v1.3.0-2013, [265,266]). Virus matches were identified using BLAST+ (version 2.10.0, [267]), spacers were blasted against the NCBI viral genomic RefSeq database [268] specifying the following parameters: word size = 8, e-value ≤ 0.01. Results were filtered for a query coverage ≥ 85% (calculated by dividing the alignment length by the query length) and allowing for a maximum number of 5 mismatches (see Supplementary Table S1), duplicate hits of a spacer to the same virus were removed.