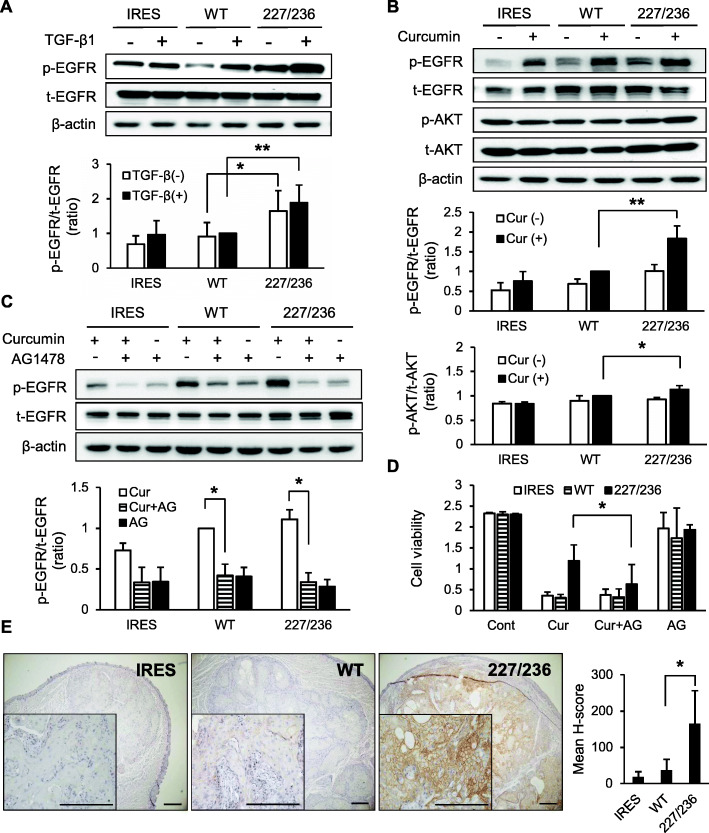
Fig. 5.
Curcumin-mediated EGFR activation in stable HSC-2 cells expressing exogenous TβRII. a. Stable HSC-2 cells were incubated in P medium containing 0.2% FBS in the presence of vehicle (−) or 10 ng/ml of TGF-β1 (+) for 24 h. EGFR protein level (t-EGFR) and phosphorylation of EGFR (p-EGFR) were detected by western blotting. Data represent mean ± standard deviation from three independent experiments (*P < 0.05, **P < 0.01). Full length immunoblots were shown in Additional file 6: Figure S6. b. Stable transfectant cells were incubated in P medium containing 0.2% FBS in the presence of vehicle (−) or 10 μ M curcumin (+) for 24 h. Activation of EGFR and AKT was detected by western blotting. t-EGFR, EGFR protein; p-EGFR, phosphorylated EGFR; t-AKT, AKT protein; p-AKT, phosphorylated AKT; Cur, curcumin. Data represent mean ± standard deviation from three independent experiments. (*P < 0.05, **P < 0.01). Full length immunoblots were shown in Additional file 7: Figure S7. c and d. Stable HSC-2 cells were incubated with 200 nM AG1478 (AG) for 30 min and treated with 10 μM curcumin (Cur) for 24 h. Activation of EGFR was determined by western blotting (c) Full length immunoblots were shown in Additional file 8: Figure S8. Cell viability was measured by MTT assay (d). Data represent mean ± standard deviation from three independent experiments (*P < 0.05). e. Immunohistochemical analysis of phosphorylated EGFR in xenograft tongue tumor tissues. Magnification, × 40 and × 200 (inset). Scale bar, 200 μm. Semiquantitative analysis of phosphorylated EGFR was performed using the H-score method. Data represent mean ± standard deviation (*P < 0.05). IRES, empty vector; WT, wild-type TβRII; 227/236, I227T/N236D TβRII