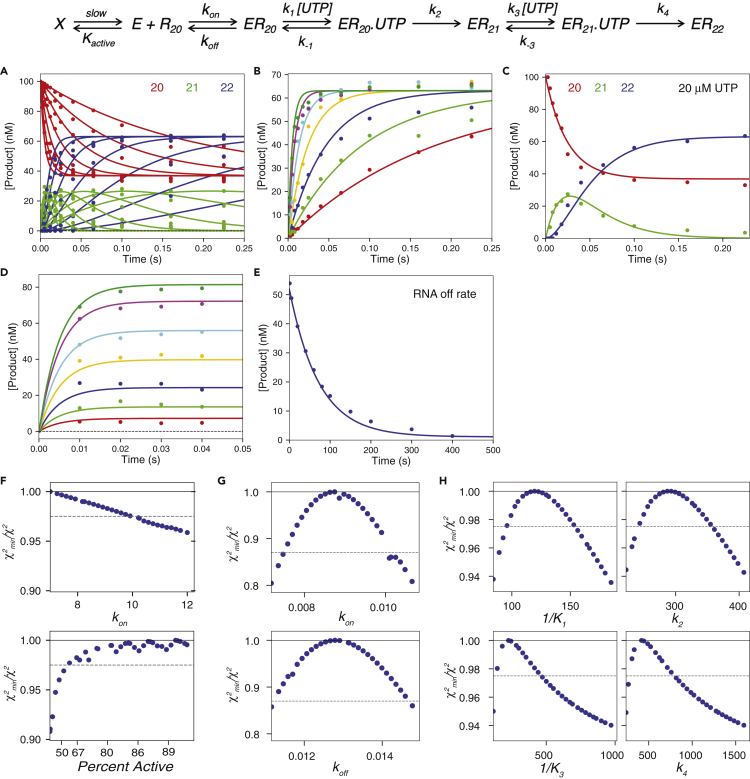
Figure 3.
SARS-CoV-2 RdRp Complex Weakly Binds an RNA Substrate and Dissociates at a Very Slow Rate
Scheme: ERn is the enzyme-RNA complex with RNA n nucleotides in length koff is the RNA dissociation rate and kon is the apparent RNA association rate. The ratio of koff/kon gives the Kd or the apparent equilibrium binding constant. Kinetic constants for UTP incorporation are labeled as in the scheme in Figure 2. Kactive is the equilibrium constant for enzyme going from the active E state to the inactive X state, which occurs during the pre-equilibration step to estimate the active enzyme concentration, as described in the Transparent Methods. Experiments in panels A–C are the same as in Figure 2.
(A) UTP incorporation, concentration dependence. Data for the 20, 21, and 22 nt RNA are shown in red, green, and blue, respectively.
(B) UTP incorporation, concentration dependence. The concentration of total product versus time is plotted for each UTP concentration.
(C) UTP incorporation at 20 μM UTP. RNA of different lengths is colored as in (A).
(D) UTP incorporation, enzyme titration. A mixture of 0.2–10 μM NSP12/7/8 complex, 20 μM NSP8, 200 nM FAM-20/40 RNA, and 5 mM Mg2+ was mixed with 150 μM UTP to start the reaction. The amount of total product formed was plotted versus time for each enzyme concentration and fit by simulation in KinTek Explorer.
(E) RNA dissociation rate experiment. A solution containing 1.25 μM NSP12/7/8 complex, 6 μM NSP8, 100 nM FAM-20/40 RNA, and 5 mM Mg2+ was allowed to equilibrate for 30 min, then mixed with 2 mg/mL heparin in the first mixing step using the RQF-3 rapid-quench flow instrument. After the designated first mixing time (shown in the figure), the reaction was mixed with 125 μM UTP (concentration after dilution) from the quench syringe, held in the exit line for 50 milliseconds, then the reaction was quenched by mixing with EDTA to a final concentration of 0.3 M in a collection tube. The time axis in the figure is the time allowed for RNA dissociation after mixing with heparin trap, before the addition of nucleotide. The best fit by simulation is shown as the solid line yielding an RNA dissociation rate of 0.013 ± 0.001 s-1
(F) Confidence contours used to estimate fraction active enzyme. Data in panels A–E were fit globally while allowing the equilibrium constant, Kactive to vary, along with kon (which compensates for variable active enzyme concentration). The data were fit to extract the percent of active enzyme and the corresponding kon values, as described in the Transparent Methods. All other rate constants were also allowed to vary, except k1 and k3 that were locked at 100 μM−1s−1. Data in the panel show that the enzyme is at least 60% active.
(G) Confidence contours to define kon and koff. Data in panels D and E were fit assuming 80% active enzyme with all other rate constants locked at their best-fit values. This analysis give kon = 0.008 ± 0.001 μM−1s−1 and koff = 0.013 ± 0.001 s−1 and Kd for RNA binding of 1.8 ± 0.2 μM.
(H) Confidence contours for UTP binding and incorporation. Date in panels A–C were fit assuming 80% active enzyme and with kon and koff locked at their best fit values. The values for k1 and k3 were locked at 100 μM−1s−1. For each contour, the dashed line shows the χ2 threshold. The smooth lines in each panel were derived from the global data fit. Rate constants derived from global data fitting, assuming 80% active enzyme, are given in Table 1.