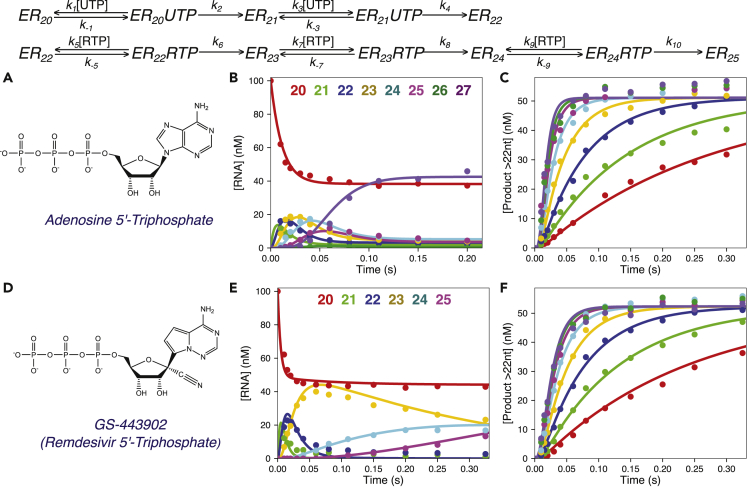
Figure 4.
Remdesivir Is Incorporated more Efficiently than ATP by the SARS-CoV-2 RdRp Complex
Scheme: kinetic pathway for UTP and ATP incorporation. Species are labeled as in Figure 2, with added steps for sequential RTP incorporations; the same sequence applies to ATP incorporation. Net rate constants k5, k7, and k9 define kcat/Km for each sequential RTP incorporation, whereas k6, k8, and k10 define kcat for each sequential RTP incorporations. For ATP the pathway is identical, but we fit the data to only define k-5 and k6 defining the kinetic parameters for the first incorporation, modeled as the sum of all products ≥23 nt in length.
(A) Chemical structure of adenosine 5′-triphosphate. Experiments: a solution containing 1.25 μM NSP12/7/8 complex, 6 μM NSP8, 100 nM FAM-20/40 RNA, and 5 mM Mg2+ was mixed with 200 μM UTP and varying concentrations of ATP (5–400 μM) to start the reaction, and products were resolved and quantified using capillary electrophoresis.
(B) Reaction progress curve for individual RNA species at 225 μM ATP. Products of primer extension from 20 to 27 nt in length are shown as a function time after mixing. Note the reaction is over in less than 0.2 s. The solid lines through the data points show the best global fit by simulation for each RNA species with an average rate of ATP incorporation around 80 s−1 at this concentration. The color code for each RNA species is given at the top of the figure.
(C) Concentration of ATP incorporation product versus time at various ATP concentrations. We plot the sum of all products after the incorporation of the first ATP, i.e., products 23–27 nt in length to define the kinetics of the first ATP incorporation. The solid lines show the best global fit by simulation for the total RNA product ≥23 nt at each ATP concentration, shown as different colors (red to purple: 5, 10, 20, 40, 80, 150, 225, and 400 μM). These data define the parameters kcat and Km for ATP incorporation as shown in Table 1.
(D) Chemical structure of GS-443902 (Remdesivir 5′-triphosphate, RTP). RTP is an adenosine analog with a 1′ cyano group and modifications to the adenine base. Note that Remdesivir contains a 3′-OH, allowing continued polymerization after its incorporation. A mixture containing 1.5 μM NSP12/7/8 complex, 6 μM NSP8, 100 nM FAM-20/40 RNA, and 5 mM Mg2+ was mixed with 150 μM UTP and varying concentrations of RTP (3.5–315 μM) to start the reaction in the quench-flow instrument. Reactions were quenched with EDTA after various reaction times and the products quantified by capillary electrophoresis.
(E) Concentration of each RNA species versus time at 315 μM RTP. The concentrations of various species, 20 nt (red), 21 nt (green), 22 nt (blue), 23 nt (yellow), 24 nt (cyan), and 25 nt (purple) are shown with the solid lines from the best global fit of the data by simulation in KinTek Explorer, which included the time course for each species at each RTP concentration (not shown).
(F) Concentration of product containing RTP versus time for various concentrations of RTP. The time dependence of the first RTP incorporation is shown as the sum of species ≥23 nt in length. The best fit by simulation is shown as the solid colored lines through the data points, with different colors for each RTP concentration (red to purple: 3.5, 7, 14, 28, 56, 112, 210, and 315 μM). The parameters kcatand Km for each Remdesivir incorporation were derived by globally fitting all of the primary data defining the formation and decay of products 23, 24, and 25 nt in length and are summarized in Table 1.