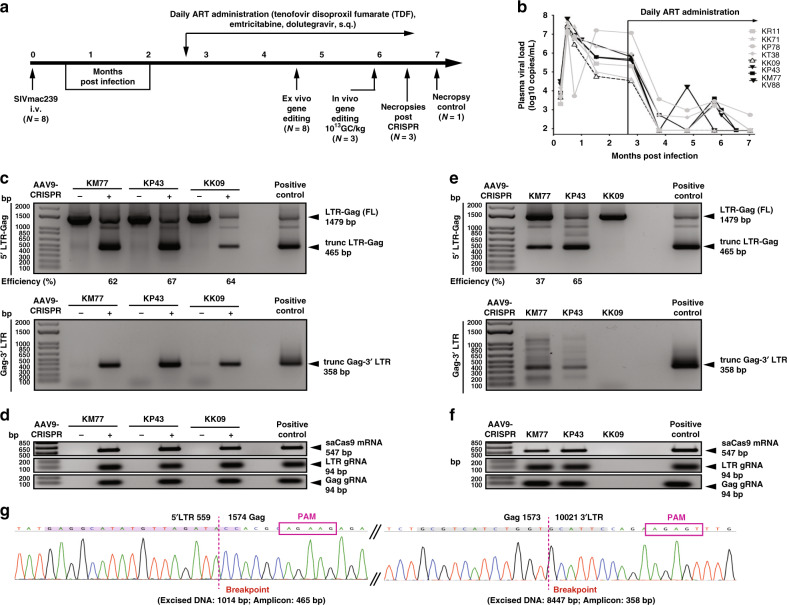
Fig. 2. Excision of SIVmac239 DNA by CRISPR-Cas9 in the blood of SIV-infected rhesus macaque monkeys.
a Chinese rhesus macaques were i.v. inoculated with SIVmac239 and given a daily ART administration of tenofovir disoproxil fumarate, emtricitabine and dolutegravir, s.q. starting at 78 days post infection. Three animals (KM77, KP43 and KV88) were given 1013 GC/kg, i.v. and 3 weeks later underwent necropsy. One animal served as the no CRISPR control (KK09). b Plasma viral loads of the SIV-infected ART-treated rhesus macaques. The threshold of the assay was 83 copies/ml. The animals in solid black symbols and lines received AAV9/CRISPR/Cas9. The open black triangle with a dotted line (KK09) was sacrificed as no AAV9/CRISPR control. The gray symbols and lines were animals that were only used in the ex vivo screening. All viral loads were done in duplicate per time point. c AAV9-mediated delivery of CRISPR/Cas9 to ex vivo PBMCs from SIVmac239-infected animals was able to excise SIV viral DNA. Truncated 5′LTR-gag (465 bp) and gag-3′LTR (358 bp) amplicons were detected in lanes with AAV-9 CRISPR/Cas9 (+) but not in the untransformed (−). The percent excision efficiency ex vivo (Efficiency (%)) shown under the PCR was calculated by quantification of the excised band (Trunc.) divided by the sum of the full-length band (FL) plus the excised bands times 100% (see Supplementary Fig. S1 for all animals). d Expression was verified by the presence of SaCas9 mRNA (547 bp), LTR and Gag gRNA scaffolds (94 bp). e Similarly, in vivo excision was confirmed in the blood of KM77 and KP43 by the PCR amplification and detection of the trunc 5′LTR to gag (465 bp) and the gag-3′LTR (358 bp) (see Supplementary Fig. S3 for KV88). The percent excision efficiency (Efficiency (%)) in vivo shown under the PCR was calculated as above. KK09 did not receive CRISPR so no efficiency was calculated. f Expression was verified by the presence of SaCas9 mRNA (547 bp), LTR and Gag gRNA scaffolds (94 bp). g Representative Sanger sequence tracings of 5′LTR-Gag (left) and Gag-3′LTR (right) CRISPR-Cas9 induced truncated SIV-specific amplicons. Target sites are highlighted in green, PAMs motifs in red, the double cleaved/end-joined site is shown as a breaking point in red. Full sequencing data are available in the source file data provided with this paper. Source data are provided as a Source Data file.