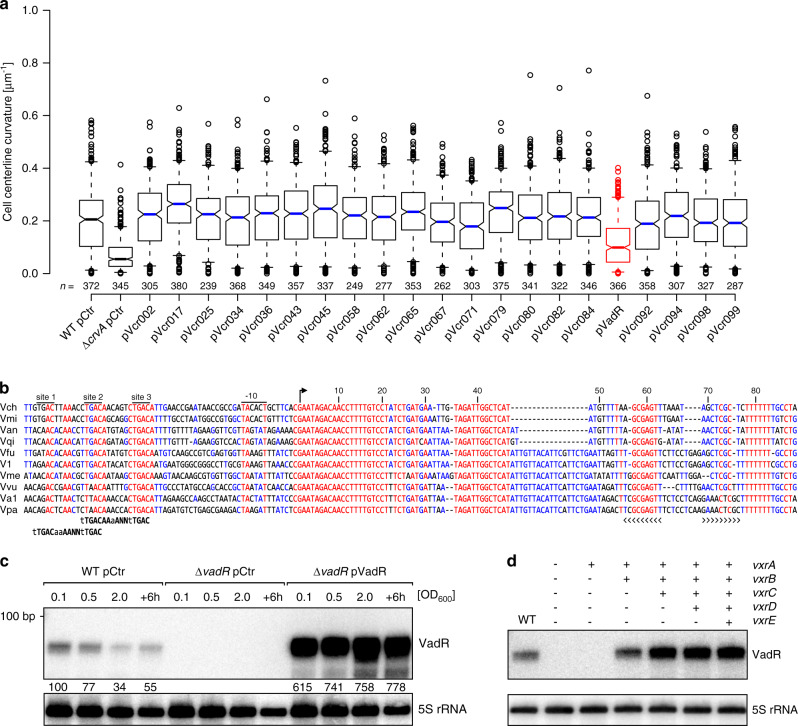
Fig. 1. The VadR sRNA alters V. cholerae cell shape and is activated by VxrB.
a Centerline curvature of V. cholerae cells expressing the indicated sRNAs (x-axis). The blue, black and red lines indicate the median, boxes represent 25th and 75th percentiles, whiskers represent 5th and 95th percentiles and notches indicate 95% confidence intervals for each median. n of each set is listed above the x-axis over three independent experiments. b Alignment of vadR and its promoter sequence from various Vibrio species (see “Methods” section “Sequence alignment” for details). The -10 box, TSS (arrow) and the Rho-independent terminator (brackets) are indicated. Putative VxrB binding sites and binding motifs (bold) are illustrated. c VadR expression throughout bacterial growth was monitored on Northern blots. V. cholerae wild-type or vadR mutant cells carrying either a control plasmid (pCtr) or a constitutive vadR overexpression plasmid (pVadR) were tested. A quantification of VadR expression relative to the wild-type (at OD600 of 0.1) is provided. d V. cholerae ΔvxrABCDE cells were complemented with various cistrons of the vxrABCDE operon and tested for VadR expression on Northern blots. Expression of the vxrABCDE fragments was driven by the inducible pBAD promoter (0.02 % l-arabinose final conc.) and exponentially growing cells were harvested (OD600 of 0.2). A V. cholerae wild-type strain harboring an empty vector served as control. The experiment was performed with three independent biological replicates (n = 3). Source data underlying panels a, c, and d are provided as a Source Data file.