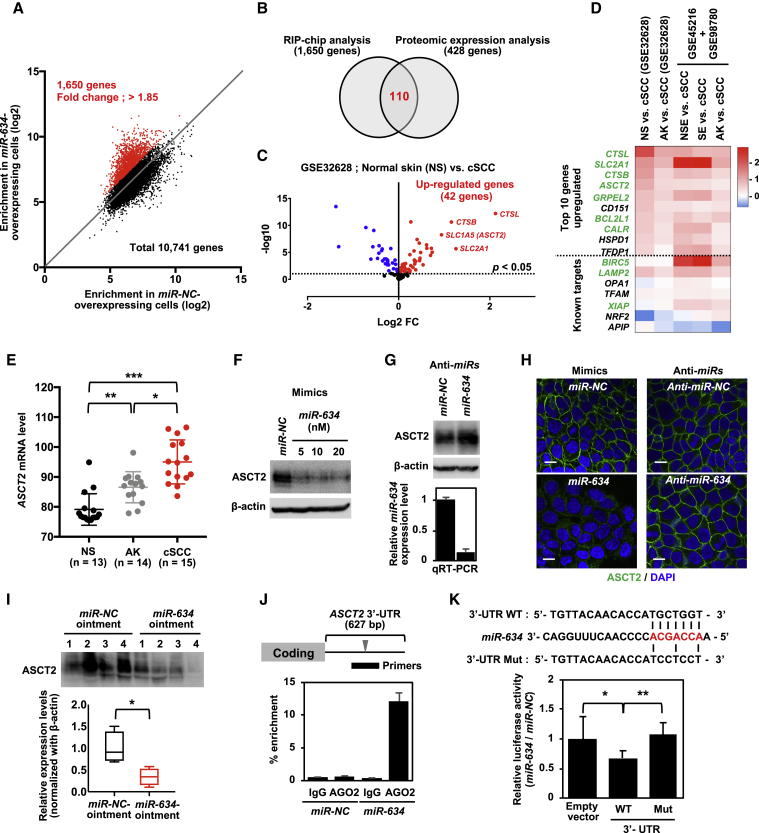
Figure 3.
Identification of ASCT2/SLC1A5 as a miR-634 Target Gene
(A) Correlation plot of enrichment (RIP/input) of 10,741 genes between miR-NC-overexpressing cells and miR-634-overexpressing cells in the RIP-chip analysis. A total of 1,650 genes were extracted as enriched in the RIP following miR-634 overexpression (red). (B) Proteomic expression analysis identified 428 genes that were downregulated at the protein level in miR-634-overexpressing cells compared with miR-NC-overexpressing cells (fold change [FC] < 1.0). Finally, 110 genes identified by both analyses were defined as directly targeted by miR-634 in A431 cells. (C) Volcano plot of log2 ratio versus p value of differentially expressed genes between primary cSCC samples (n = 15) and normal skin (NS; n = 13) on an expression array dataset (GEO: GSE32628). Significantly upregulated or downregulated genes were indicated as red or blue dots, respectively. The dotted line indicates an adjusted p value threshold of 0.05. (D) Heatmap of log2 FCs of cSCC versus non-cancerous tissues from GEO: GSE32628 and a combined dataset of GEO: GSE98780 and GSE45216. AK, actinic keratosis; NSE, non-sun-exposed NS; SE, sun-exposed NS. Significantly upregulated genes in four out of five comparisons are highlighted in green. Within the heatmap, red color indicates the upregulation in cSCC compared to non-cancerous tissues, and the blue color indicates the downregulation in cSCC. (E) Expression level of ASCT2 in NS (n = 13, black), AK (n = 14, gray), and cSCC (n = 15, red) from GEO: GSE32628. p values were calculated using the two-sided Student’s t test (∗p = 0.014 for cSCC versus AK, ∗∗p = 0.0006 for NS versus AK, and ∗∗∗p < 0.0001 for cSCC versus NS). (F) Western blotting analysis of ASCT2 in miR-634-overexpressing cells. After 3 days of transfection, cell lysates were prepared, separated by SDS-PAGE, and immunoreacted with the indicated antibodies. (G) Western blotting analysis of ASCT2 and expression analysis of miR-634 in miR-634-inhibited cells. A431 cells were transfected with 100 nM anti-miR-NC or anti-miR-634. Upper panel: after 3 days of transfection, cell lysates were prepared, separated by SDS-PAGE, and immunoreacted with the indicated antibodies. Lower panel: qRT-PCR analysis of miR-634 expression. Error bars indicate the SD. Data are presented as mean ± SD. (H) Immunofluorescence analysis of ASCT2. After 3 days of transfection, cells were fixed, blocked, and incubated with anti-ASCT2 antibodies. Images were obtained by confocal fluorescence microscopy. Scale bars, 10 μm. (I) Western blotting analysis of resected tumors. The cell lysates described in Figure 1I were separated by SDS-PAGE and immunoreacted with anti-ASCT2 antibodies. The expression levels of target genes in tumors treated with miR-634 ointment relative to those treated with miR-NC ointment are indicated as boxplots. p values were calculated using the two-sided Student’s t test (∗p = 0.019). (J) RIP-PCR analysis of the ASCT2 gene. Upper panel: a putative miR-634 binding site within the 3′ UTR of the ASCT2 gene. Lower panel: enrichment relative to IgG immunoprecipitation in miR-NC-overexpressing cells. Data are presented as mean ± SD. (K) Luciferase assays using reporter plasmids. The luciferase activity in miR-634-transfected cells relative to that in miR-NC-transfected cells is indicated on the vertical axis. Error bars indicate the SD. Data are presented as mean ± SD. p values were calculated using one-way ANOVA (∗p = 0.024 to empty vector, ∗∗p = 0.0046 to Mut vector).