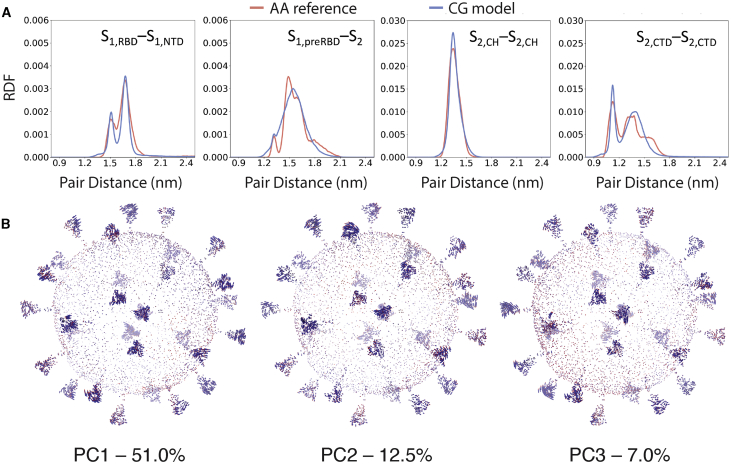
Figure 4.
Analysis of the CG MD simulations of the SARS-CoV-2 virion. (A) RDFs showing the comparison between mapped AA reference statistics and the CG spike model during the MD simulations are given. The measured RDFs are for CG particles that were mapped from the following AA residues of 1) S1,RBD [S459-D467] and S1,NTD [W104-L118], 2) S1,preRBD [E309-R319] and S2 [A852-L861], 3) S2,CH [A1015-K1028], and 4) S2,CTD [Y1215-V1228]. (B) Principal modes of motion of the SARS-CoV-2 virion computed from the CG MD simulation are shown. Arrows are colored from blue to red, indicating the direction of movement (see Videos S2, S3, and S4 for PC1–3, respectively). The first principal component (PC1) accounts for 51% of the total variation observed during simulation, whereas the second (PC2) and third (PC3) account for 12.5 and 7%, respectively. To see this figure in color, go online.