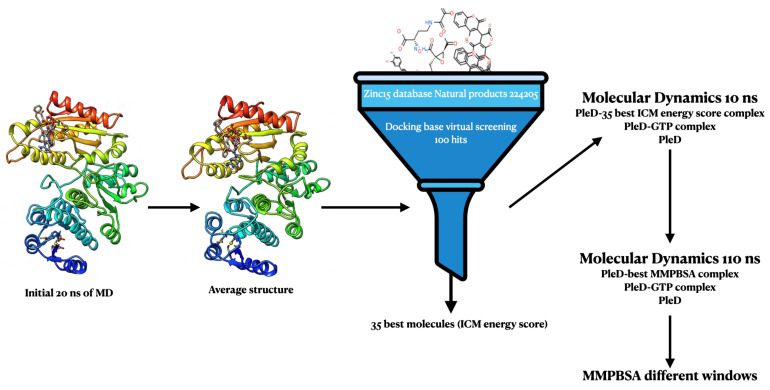
Figure 1.
Summary of procedures used for the hit identification. An initial 20 ns of molecular dynamics (MD) was performed with the PleD protein (inactive form) in complex with guanosine-5-triphosphate (GTP) molecule and Mg ions, by following the crystal structure of the active form 2V0N to coupling the GTP at the catalytic site. From an equilibrated trajectory starting at 1.2 ns, an average structure was produced using chimera software [26]. A library of about 224,205 natural products compounds was obtained from ZINC15 database. Bond orders, tautomeric forms, stereochemistry, hydrogen atoms, and protonation states were assigned automatically by the ICM package, with default parameters for each compound to prepare the ligands for docking. Ligands were docked using virtual screening scheme from ICM using the PleD protein average structure obtained by the above procedure. The GTP geometric space in the protein was used as a reference for virtual screening (VS) procedure and ICM energy scoring function was used for ranking it. From the 100 hits, 35 with the best ICM energy score were selected for 10 ns of MD simulations. For each molecule, MM/PBSA was computed using 200 frames from the last 2 ns of each MD. Finally, with the best complex (Ligand4–PleD), ranked from MM/PBSA binding free energy, a 110 ns of MD was done. This was also done for the GTP–PleD protein complex and PleD protein. From the above three trajectories, MM/PBSA was computed again at different windows starting from 20 ns until 60 ns using 200 frames for each window.