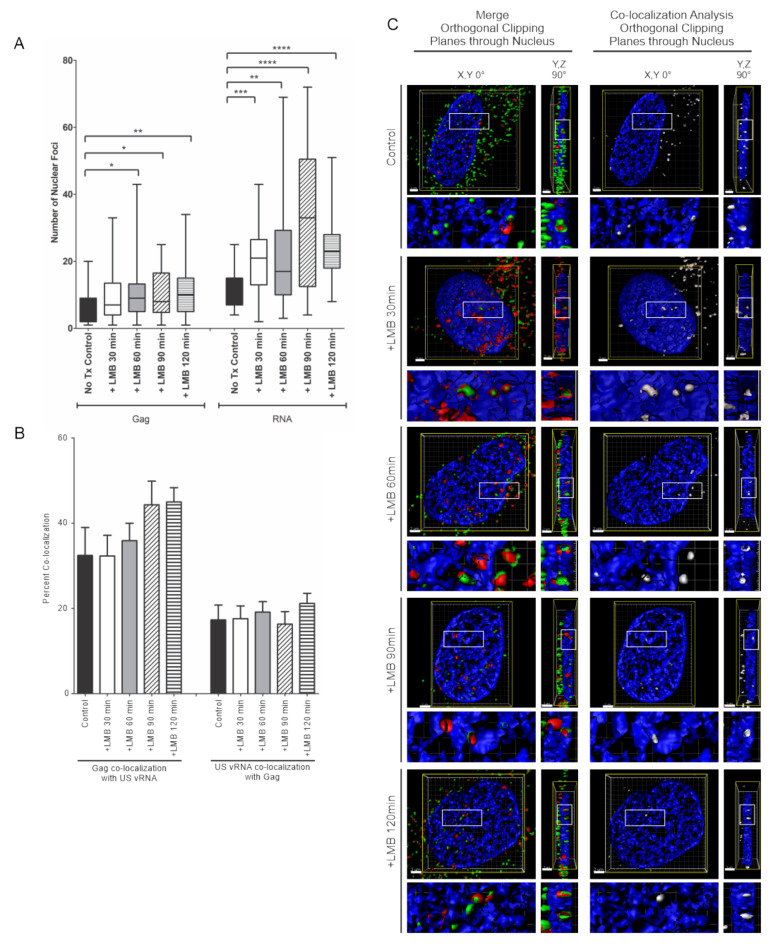
Figure 6.
Inhibiting CRM1-mediated nuclear export in HeLa HIV.Gag-GFP rtTA cells increased HIV-1 Gag and USvRNA nuclear foci but did not affect the degree of co-localization between HIV-1 Gag and USvRNA. (A) Quantitative analysis depicting the mean number of HIV-1 Gag-GFP and USvRNA nuclear foci in dox-induced HeLa HIV.Gag-GFP rtTA cells following LMB treatment compared to the untreated control cells. (B) Quantitative analysis of dox-induced HIV-1 Gag-GFP rtTA cells demonstrating the mean percent of nuclear HIV-1 Gag co-localization with USvRNA and USvRNA co-localization with HIV-1 Gag following treatment with LMB for 30, 60, 90, or 120 min prior to fixation compared to the untreated control. (error bars = standard error of the mean, statistical significances: * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001). (C) 3D surface renderings of dox-induced HeLa HIV.Gag-GFP rtTA cells depicting the co-localization (shown in white) of HIV-1 Gag-GFP (green) and USvRNA (red, detected by FISH) foci within the nucleus (DAPI, blue) following treatment with LMB. 3D surface renderings were subjected to an orthogonal clipping plane bisected in the X, Y and Y, Z planes to show the co-localization of foci within the nucleus. The white rectangle represents the region of interest enlarged in the panels below.